5WEO
 
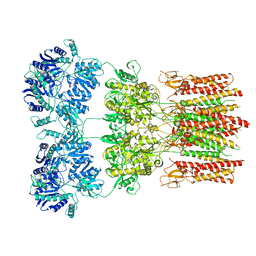 | | Activated GluA2 complex bound to glutamate, cyclothiazide, and STZ in digitonin | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit chimera | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
5XAA
 
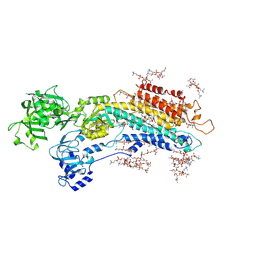 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of P21212 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA8
 
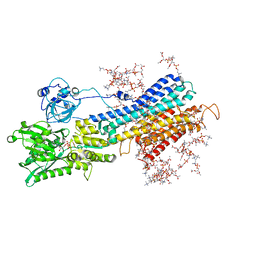 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-ALF4-ADP-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA9
 
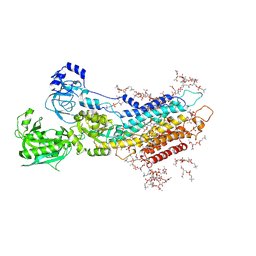 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of C2 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA7
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XAB
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2(TG) crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
7QHB
 
 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
5X8S
 
 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Ursolic acid. | | Descriptor: | Nuclear receptor ROR-gamma, Ursolic acid | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
5YLV
 
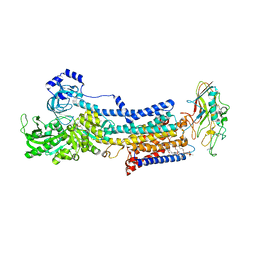 | | Crystal structure of the gastric proton pump complexed with SCH28080 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(2-methyl-8-phenylmethoxy-imidazo[1,2-a]pyridin-3-yl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79977775 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
5YLU
 
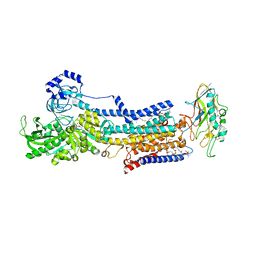 | | Crystal structure of the gastric proton pump complexed with vonoprazan | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[5-(2-fluorophenyl)-1-pyridin-3-ylsulfonyl-pyrrol-3-yl]-~{N}-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79988956 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
5NCQ
 
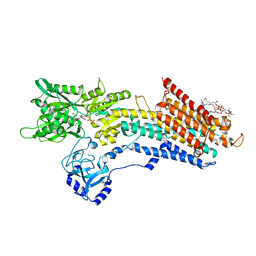 | | Structure of the (SR) Ca2+-ATPase bound to a Tetrahydrocarbazole and TNP-ATP | | Descriptor: | (1~{S})-~{N}-[(4-bromophenyl)methyl]-7-(trifluoromethyloxy)-2,3,4,9-tetrahydro-1~{H}-carbazol-1-amine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, ... | | Authors: | Bublitz, M, Kjellerup, L, O'Hanlon Cohrt, K, Gordon, S, Mortensen, A.L, Clausen, J.D, Pallin, D, Hansen, J.B, Brown, W.D, Fuglsang, A, Winther, A.-M.L. | | Deposit date: | 2017-03-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity.
PLoS ONE, 13, 2018
|
|
7RQW
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an open state, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
5LRX
 
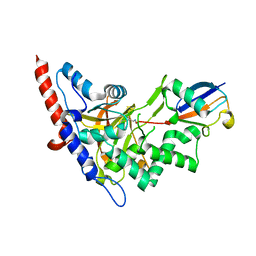 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
4UEL
 
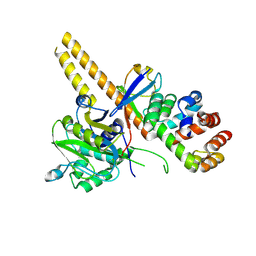 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | Descriptor: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
6AAO
 
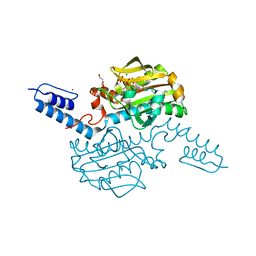 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
4UU1
 
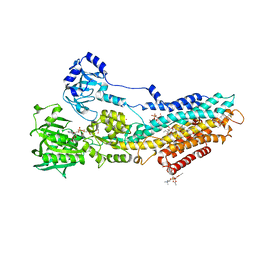 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF DOPC | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
6U8A
 
 | |
3IRX
 
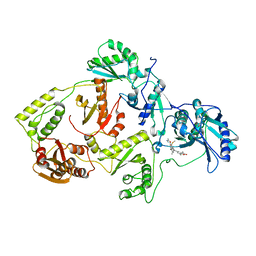 | | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the Non-nucleoside RT Inhibitor (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate. | | Descriptor: | (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methy lbenzothioate, Reverse transcriptase, Reverse transcriptase/ribonuclease H | | Authors: | Ho, W.C, Arnold, E. | | Deposit date: | 2009-08-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the
alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor (E)-S-Methyl
5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate.
J.Med.Chem., 52, 2009
|
|
6U88
 
 | |
6UP0
 
 | |
6V4H
 
 | | Crystal Structure Analysis of Zebra Fish MDMX | | Descriptor: | Protein Mdm4, Stapled Peptide LSQETF(0EH)DLWKLL(MK8)EN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
6V4F
 
 | | Crystal Structure Analysis of Zebra Fish MDMX | | Descriptor: | Protein Mdm4, SULFATE ION, Stapled Peptide LSQETF(0EH)DLWKLE(MK8)EN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
3ZNH
 
 | | Crimean Congo Hemorrhagic Fever Virus OTU domain in complex with ubiquitin-propargyl. | | Descriptor: | POLYUBIQUITIN-B, UBIQUITIN THIOESTERASE | | Authors: | Ekkebus, R, vanKasteren, S.I, Kulathu, Y, Scholten, A, Berlin, I, deJong, A, Goerdayal, G, Neefjes, J, Heck, A.J.R, Komander, D, Ovaa, H. | | Deposit date: | 2013-02-14 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On Terminal Alkynes that Can React with Active-Site Cysteine Nucleophiles in Proteases.
J.Am.Chem.Soc., 135, 2013
|
|
3K52
 
 | |
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
