7KEE
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7DMB
 
 | |
7JU2
 
 | | Crystal structure of the monomeric ETV6 PNT domain | | Descriptor: | FORMIC ACID, Transcription factor ETV6 | | Authors: | Gerak, C.A.N, Kolesnikov, M, Murphy, M.E.P, McIntosh, L.P. | | Deposit date: | 2020-08-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85002184 Å) | | Cite: | Biophysical characterization of the ETV6 PNT domain polymerization interfaces.
J.Biol.Chem., 296, 2021
|
|
4P1E
 
 | | Crystal structure of a trap periplasmic solute binding protein from escherichia fergusonii (efer_1530), target EFI-510119, apo open structure, phased with iodide | | Descriptor: | IODIDE ION, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCZ
 
 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | Authors: | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LHB
 
 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | Descriptor: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | Authors: | Judge, R.A, Salem, A.H. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
7KJ9
 
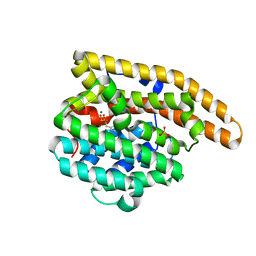 | | Wild-type epi-isozizaene synthase: complex with 3 Mg2+ and risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
7KMV
 
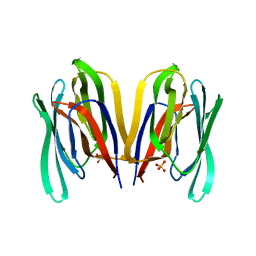 | | Structure of Malaysian Banana Lectin F84T | | Descriptor: | Jacalin-type lectin domain-containing protein, SULFATE ION | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
7L5E
 
 | |
7KJE
 
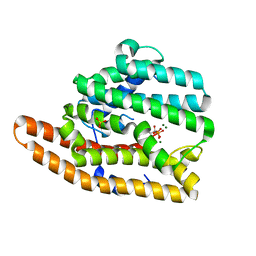 | | F96S epi-isozizaene synthase: complex with 3 Mg2+ and neridronate | | Descriptor: | (6-azanyl-1-oxidanyl-1-phosphono-hexyl)phosphonic acid, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
7LWC
 
 | | Goat beta-lactoglobulin mutant Q59A | | Descriptor: | Beta-lactoglobulin | | Authors: | Munoz, D.A, Dobson, R.C.J. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering food proteins for improved digestibility and lower allergenicity: a case study using caprine beta-lactoglobulin
To Be Published
|
|
7LTG
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH APICIDIN | | Descriptor: | (3S,6S,9S,15aR)-9-[(2S)-butan-2-yl]-3-(6,6-dihydroxyoctyl)-6-[(1-methoxy-1H-indol-3-yl)methyl]octahydro-2H-pyrido[1,2-a][1,4,7,10]tetraazacyclododecine-1,4,7,10(3H,12H)-tetrone, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Beshore, D.C. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction.
Acs Med.Chem.Lett., 12, 2021
|
|
7LTK
 
 | |
7LTL
 
 | |
7LHZ
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
7LXV
 
 | | Structure of human 20S proteasome with bound MPI-5 | | Descriptor: | N-[(1R)-2-([1,1'-biphenyl]-4-yl)-1-boronoethyl]-1-methyl-L-prolinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXT
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound bortezomib | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Morton, C.J, Metcalfe, R.D, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXU
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound MPI-5 | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Xie, S.C, Liu, B, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LEW
 
 | | Crystal structure of UBE2G2 in complex with the UBE2G2-binding region of AUP1 | | Descriptor: | Lipid droplet-regulating VLDL assembly factor AUP1, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Liang, Y.-H, Smith, C.E, Tsai, Y.C, Weissman, A.M, Ji, X. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | A structurally conserved site in AUP1 binds the E2 enzyme UBE2G2 and is essential for ER-associated degradation.
Plos Biol., 19, 2021
|
|
4O2D
 
 | |
7C61
 
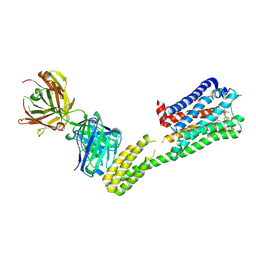 | | Crystal structure of 5-HT1B-BRIL and SRP2070_Fab complex | | Descriptor: | 5-hydroxytryptamine receptor 1B,Soluble cytochrome b562,5-hydroxytryptamine receptor 1B, Ergotamine, IGG HEAVY CHAIN, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
3O9W
 
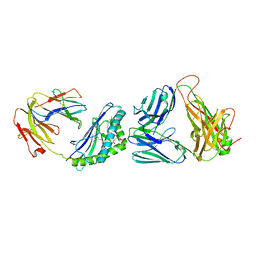 | | Recognition of a Glycolipid Antigen by the iNKT Cell TCR | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
4ILC
 
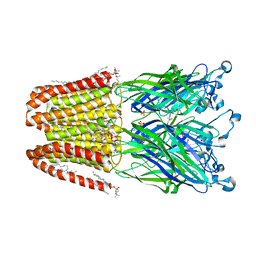 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
