6TME
 
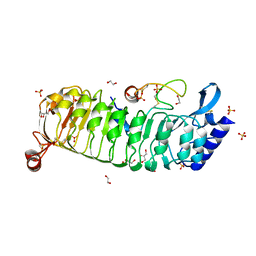 | | Monomeric LRX8 in complex with RALF4. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7WR3
 
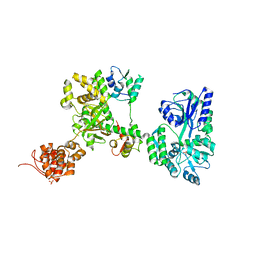 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T6L
 
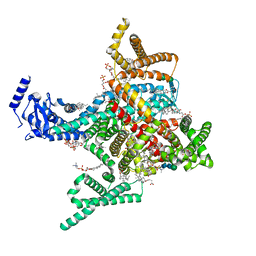 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
2INV
 
 | |
7CA5
 
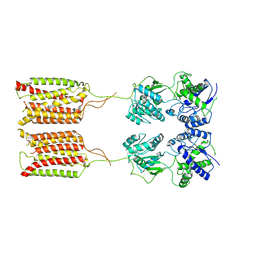 | | Cryo-EM structure of human GABA(B) receptor in apo state | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7WR5
 
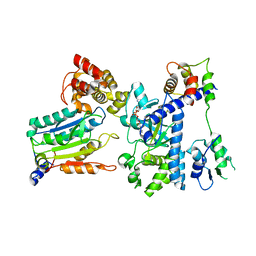 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LXC
 
 | |
6NPA
 
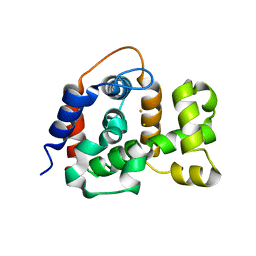 | | X-ray crystal structure of TmpB, (R)-1-hydroxy-2-trimethylaminoethylphosphonate oxygenase, with (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, FE (III) ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6MU1
 
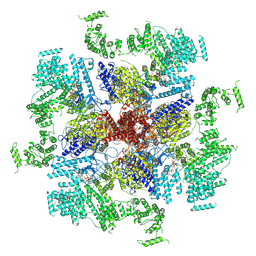 | | Structure of full-length IP3R1 channel bound with Adenophostin A | | Descriptor: | Adenophostin A, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
7WSX
 
 | |
5KRS
 
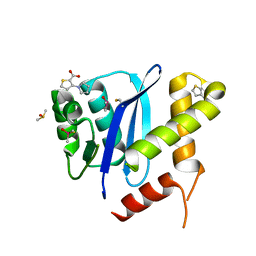 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
6N5W
 
 | |
1KYT
 
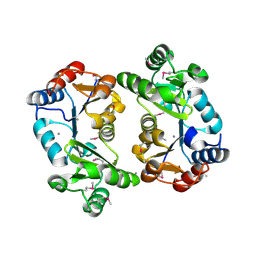 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
6BUT
 
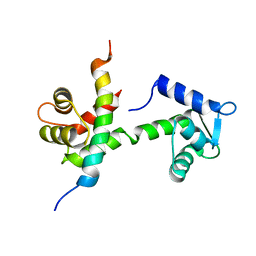 | |
1L3N
 
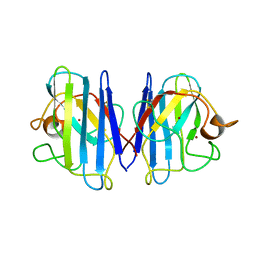 | | The Solution Structure of Reduced Dimeric Copper Zinc SOD: the Structural Effects of Dimerization | | Descriptor: | COPPER (I) ION, ZINC ION, superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-05-08 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization
Eur.J.Biochem., 269, 2002
|
|
6C6Q
 
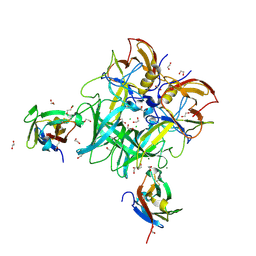 | |
1L1N
 
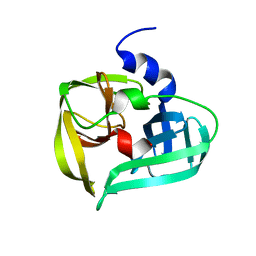 | | POLIOVIRUS 3C PROTEINASE | | Descriptor: | Genome polyprotein: Picornain 3C | | Authors: | Mosimann, S.C, Chernaia, M.M, Sia, S, Plotch, S, James, M.N.G. | | Deposit date: | 2002-02-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined X-ray crystallographic structure of the poliovirus 3C gene product.
J.Mol.Biol., 273, 1997
|
|
8HJT
 
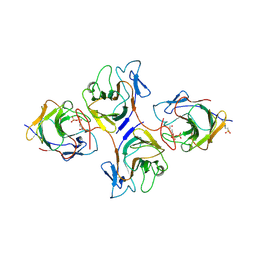 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6C9D
 
 | |
5M54
 
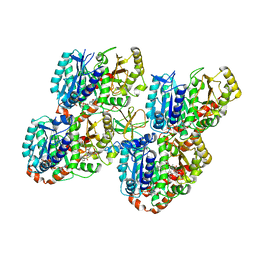 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6TO6
 
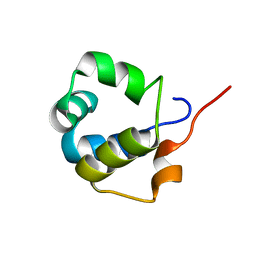 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2KS0
 
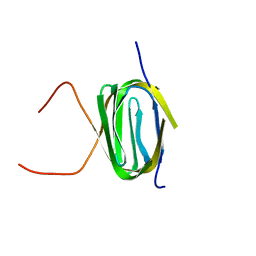 | | Solution NMR structure of the Q251Q8_DESHY(21-82) protein from Desulfitobacterium Hafniense, Northeast Structural Genomics Consortium Target DhR8C | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
6NPB
 
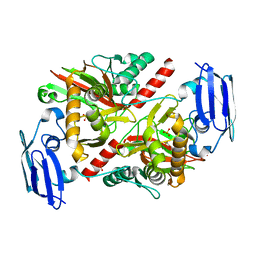 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, SULFATE ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
7Y7T
 
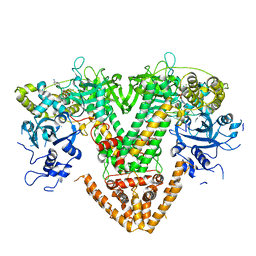 | | QDE-1 in complex with 12nt DNA template, ATP and 3'-dGTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*G*AP*GP*AP*CP*CP*TP*TP*TP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
8FVJ
 
 | | Dimeric form of HIV-1 Vif in complex with human CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Elongin-B, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
