8VZ1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-409 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(6-methoxy-3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VYT
 
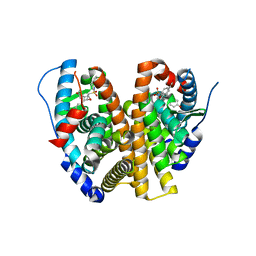 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-411 | | Descriptor: | 4,4'-[(1R,4R,5S)-5-(2,3-dihydro-1H-indole-1-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZP
 
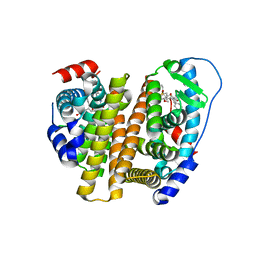 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-403 | | Descriptor: | (1S,2R,4S)-N-(2-hydroxyethyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VYX
 
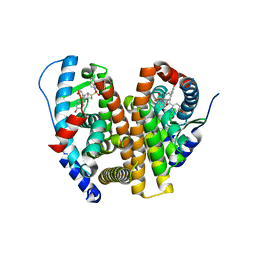 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-410 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZQ
 
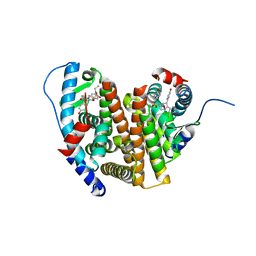 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-406 | | Descriptor: | (1S,2R,4S)-N-(2-fluoro-2-methylpropyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NICKEL (II) ION | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W03
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-1154 | | Descriptor: | (1S,2R,4S)-N-(4-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6W2G
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A in Monoclinic Unit Cell | | Descriptor: | 1,2-ETHANEDIOL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
6W2H
 
 | | Crystal Structure of the Internal UBA Domain of HHR23A | | Descriptor: | SULFATE ION, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Residual Structure in the Denatured State of the Fast-Folding UBA(1) Domain from the Human DNA Excision Repair Protein HHR23A.
Biochemistry, 61, 2022
|
|
6W2I
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A | | Descriptor: | GLYCEROL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
2J8Z
 
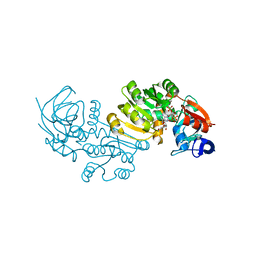 | | Crystal Structure of human P53 inducible oxidoreductase (TP53I3,PIG3) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE | | Authors: | Pike, A.C.W, Shafqat, N, Debreczeni, J, Johansson, C, Haroniti, A, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, von Delft, F, Porte, S, Fita, I, Pares, J, Pares, X, Oppermann, U. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure and Enzymatic Function of Proapoptotic Human P53-Inducible Quinone Oxidoreductase Pig3.
J.Biol.Chem., 284, 2009
|
|
8F35
 
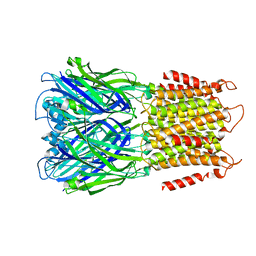 | | Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F33
 
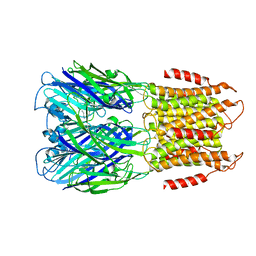 | | ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F34
 
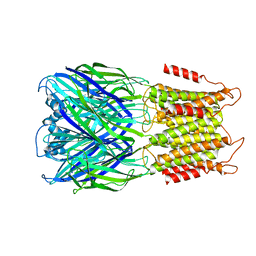 | | ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F32
 
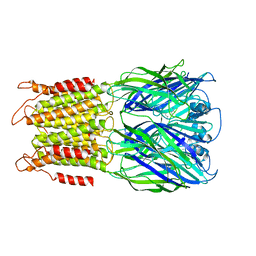 | | ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8FY5
 
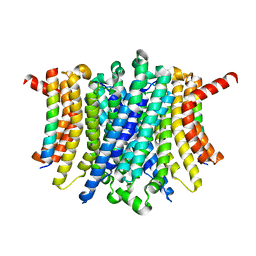 | | Human TMEM175-LAMP1 full-length complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
8FYF
 
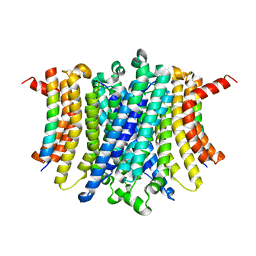 | | Human TMEM175-LAMP1 transmembrane domain only complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
5SW8
 
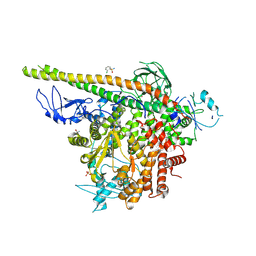 | | Crystal structure of PI3Kalpha in complex with fragments 7 and 11 | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, 2H-indazol-5-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
3VBD
 
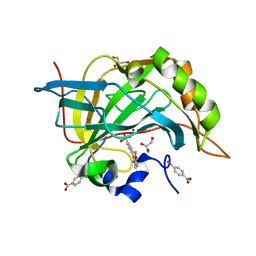 | | Complex of human carbonic anhydrase II with 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide | | Descriptor: | 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2012-01-02 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
4HLA
 
 | | Crystal structure of wild type HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
5SXK
 
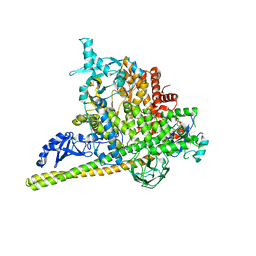 | | Crystal Structure of PI3Kalpha in complex with fragment 18 | | Descriptor: | 2-methylbenzene-1,3-diamine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
4I7J
 
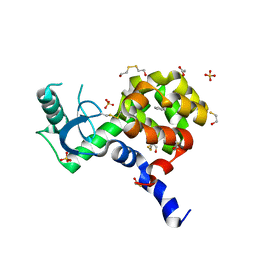 | | T4 Lysozyme L99A/M102H with benzene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BENZENE, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I8W
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL007 | | Descriptor: | 4-{[(2R,3S)-3-({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yloxy]carbonyl}amino)-2-hydroxy-4-phenylbutyl](2-methylpropyl)sulfamoyl}benzoic acid, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
2IWA
 
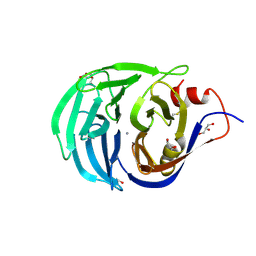 | | Unbound glutaminyl cyclotransferase from Carica papaya. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLUTAMINE CYCLOTRANSFERASE, ... | | Authors: | Guevara, T, Mallorqui-Fernandez, N, Garcia-Castellanos, R, Petersen, G.E, Lauritzen, C, Pedersen, J, Arnau, J, Gomis-Ruth, F.X, Sola, M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Papaya Glutamine Cyclotransferase Shows a Singular Five-Fold Beta-Propeller Architecture that Suggests a Novel Reaction Mechanism.
Biol.Chem., 387, 2006
|
|
8CRL
 
 | | Crystal structure of LplA1 in complex with the inhibitor C3 (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRI
 
 | | Crystal structure of LplA1 in complex with lipoic acid (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LIPOIC ACID, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
