3MWI
 
 | | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine | | Descriptor: | 5-nitro-1H-indole-2-carboximidamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Yu, H.Y, Yuan, C, Huang, Z.X, Huang, M.D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine
To be Published
|
|
4CN9
 
 | | structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus with zinc occupied MIDAS motif | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PROXIMAL THREAD MATRIX PROTEIN 1, ... | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
6E8M
 
 | |
3N53
 
 | |
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CXN
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form I | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MU1
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MVS
 
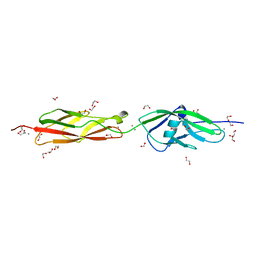 | | Structure of the N-terminus of Cadherin 23 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-23 | | Authors: | Clark, P, Joseph, J.S, Kolatkar, A.R. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6E6F
 
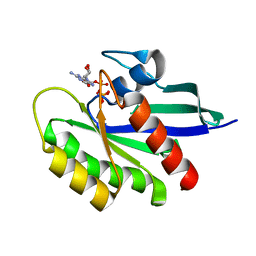 | | KRAS G13D bound to GppNHp (K13GNP) | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
3MUV
 
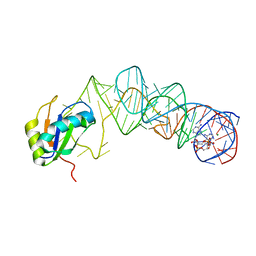 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
6E6G
 
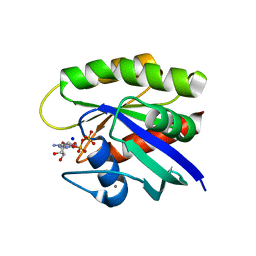 | | KRAS G13D bound to GDP (K13GDP) | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
3MWR
 
 | |
4D6A
 
 | |
6E7M
 
 | |
4D7S
 
 | | Structure of the SthK Carboxy-Terminal Region in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, STHK_CNBD_CGMP | | Authors: | Kesters, D, Brams, M, Nys, M, Wijckmans, E, Spurny, R, Voets, T, Tytgat, J, Ulens, C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels.
Plos One, 10, 2015
|
|
3MZR
 
 | | RNase crystals grown in loops/micromounts | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Mathews, I.I. | | Deposit date: | 2010-05-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
4COH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the sulfonamide derivative of the 4-aminopyridyl-based inhibitor | | Descriptor: | 2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]-4-(4-thiophen-2-ylsulfonylpiperazin-1-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Vieira, D.F, Choi, J.Y, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Expanding the Binding Envelope of Cyp51 Inhibitors Targeting Trypanosoma Cruzi with 4-Aminopyridyl-Based Sulfonamide Derivatives
Chembiochem, 15, 2014
|
|
3N3E
 
 | | Zebrafish AlphaA crystallin | | Descriptor: | Alpha A crystallin | | Authors: | Laganowsky, A, Eisenberg, D. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Non-3D domain swapped crystal structure of truncated zebrafish alphaA crystallin.
Protein Sci., 19, 2010
|
|
3N4L
 
 | | BACE-1 in complex with ELN380842 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-({1-[3-(1H-pyrazol-1-yl)phenyl]cyclohexyl}amino)propyl]acetamide | | Authors: | Yao, N.H. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6EOE
 
 | | Crystal structure of AMPylated GRP78 with nucleotide | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
4D78
 
 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
4CNB
 
 | | Structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus - Crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, PROXIMAL THREAD MATRIX PROTEIN 1, SULFATE ION | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
