8PDY
 
 | | E. coli RNA polymerase paused at ops site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PH9
 
 | | E. coli RNA polymerase paused at ops site (non-complementary scaffold) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
2Q66
 
 | | Structure of Yeast Poly(A) Polymerase with ATP and oligo(A) | | Descriptor: | 1,2-ETHANEDIOL, 5'-R(P*AP*AP*AP*AP*A)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bohm, A, Balbo, P. | | Deposit date: | 2007-06-04 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis.
Structure, 15, 2007
|
|
5N9J
 
 | | Core Mediator of transcriptional regulation | | Descriptor: | Mediator Complex Subunit 9, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Nozawa, K, Schneider, T.R, Cramer, P. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Core Mediator structure at 3.4 Angstrom extends model of transcription initiation complex.
Nature, 545, 2017
|
|
6CCE
 
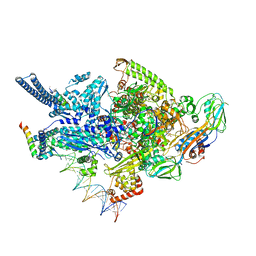 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (57-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
5ZYP
 
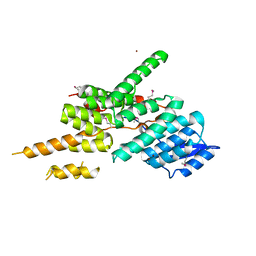 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
8GWE
 
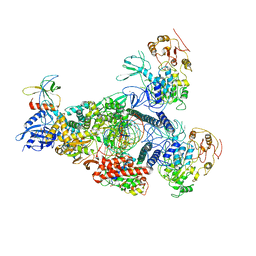 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
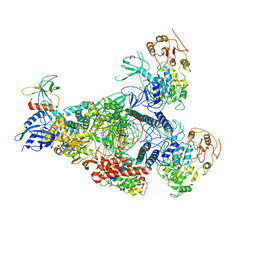 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
1UON
 
 | | REOVIRUS POLYMERASE LAMBDA-3 LOCALIZED BY ELECTRON CRYOMICROSCOPY OF VIRIONS AT 7.6-A RESOLUTION | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*GP*GP*GP*GP*GP*)-3', 5'-R(*UP*AP*GP*CP*CP*CP*CP*CP*)-3', ... | | Authors: | Zhang, X, Walker, S.B, Chipman, P.R, Nibert, M.L, Baker, T.S. | | Deposit date: | 2003-09-21 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Reovirus Polymerase Lambda3 Localized by Cryo-Electron Microscopy of Virions at a Resolution of 7.6 A
Nat.Struct.Biol., 10, 2003
|
|
5ZAM
 
 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
8GWO
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
9F6D
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (open conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9F6F
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9F6E
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QJ7
 
 | |
9GUK
 
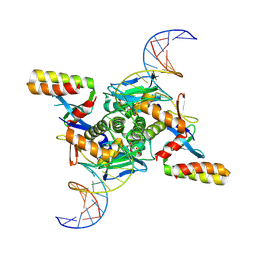 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus in complex with its transcriptional co- activator PipX and its target DNA (Crystal I) | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (30-MER), Global nitrogen regulator, ... | | Authors: | Forcada-Nadal, A, Llacer, J.L, Rubio, V. | | Deposit date: | 2024-09-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures of the cyanobacterial nitrogen regulators NtcA and PipX complexed to DNA shed light on DNA binding by NtcA and implicate PipX in the recruitment of RNA polymerase.
Nucleic Acids Res., 53, 2025
|
|
3VU7
 
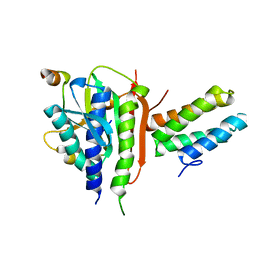 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
4OAV
 
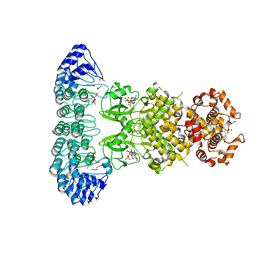 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | Descriptor: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
4OAU
 
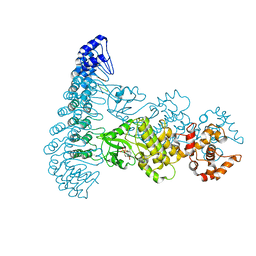 | | Complete human RNase L in complex with biological activators. | | Descriptor: | 2-5A-dependent ribonuclease, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
3J7P
 
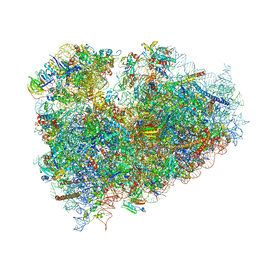 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
5VO8
 
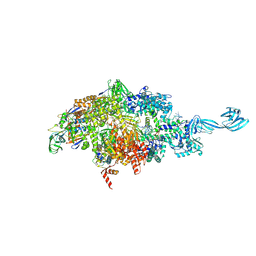 | | X-ray crystal structure of a bacterial reiterative transcription complex of pyrG promoter | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*CP*TP*GP*AP*TP*GP*CP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*GP*CP*AP*TP*CP*AP*GP*AP*GP*CP*CP*CP*AP*AP*AP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Murakami, K.S, Shin, Y, Turnbough Jr, C.L, Molodtsov, V. | | Deposit date: | 2017-05-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ZAL
 
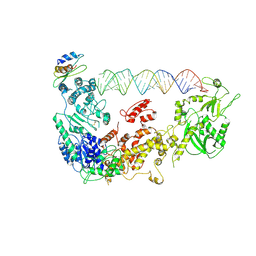 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
3C66
 
 | | Yeast poly(A) polymerase in complex with Fip1 residues 80-105 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Poly(A) polymerase, ... | | Authors: | Bohm, A, Meinke, G. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein.
Biochemistry, 47, 2008
|
|
