3N8Y
 
 | | Structure of Aspirin Acetylated Cyclooxygenase-1 in Complex with Diclofenac | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
4HFM
 
 | |
3N8Z
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
4C1K
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOHEPTONATE ALDOLASE, CADMIUM ION, CARBONATE ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Destabilization of the Homotetrameric Assembly of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase from the Hyperthermophile Pyrococcus Furiosus Enhances Enzymatic Activity
J.Mol.Biol., 426, 2014
|
|
4GVP
 
 | |
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXR
 
 | | Structure of ATP bound RpMatB-BxBclM chimera B3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, GLYCEROL, ... | | Authors: | Rank, K.C, Crosby, H.A, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Substrate Specificity of the Rhodopseudomonas palustris Protein Acetyltransferase RpPat: IDENTIFICATION OF A LOOP CRITICAL FOR RECOGNITION BY RpPat.
J.Biol.Chem., 287, 2012
|
|
1NLN
 
 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
3S34
 
 | | Structure of the 1121B Fab fragment | | Descriptor: | 1121B Fab heavy chain, 1121B Fab light chain, PHOSPHATE ION | | Authors: | Franklin, M.C. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Function of Two Anti-VEGF Receptor 2 Antibodies.
Structure, 19, 2011
|
|
2VGY
 
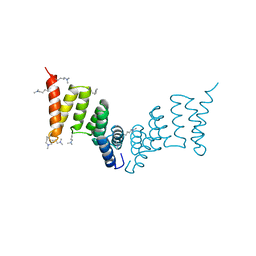 | | Crystal structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD (alternative dimer) | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
6QJH
 
 | | Cryo-EM structure of heparin-induced 2N4R tau snake filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
3TIF
 
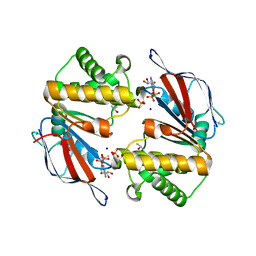 | |
1NZ0
 
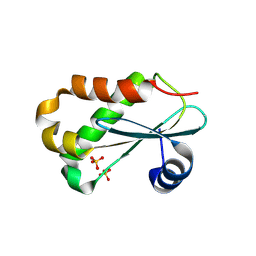 | | RNASE P PROTEIN FROM THERMOTOGA MARITIMA | | Descriptor: | Ribonuclease P protein component, SULFATE ION | | Authors: | Kazantsev, A.V, Krivenko, A.A, Harrington, D.J, Carter, R.J, Holbrook, S.R, Adams, P.D, Pace, N.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of RNase P protein from Thermotoga maritima.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4EZM
 
 | | Crystal structure of the human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 | | Descriptor: | Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, alpha-D-mannopyranose, ... | | Authors: | Dhaliwal, B, Yuan, D, Sutton, B.J. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor Fc{varepsilon}RI.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4N2S
 
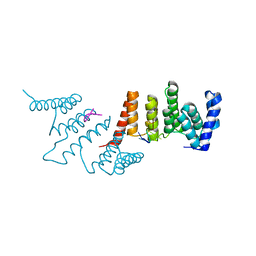 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | Descriptor: | THA8 RNA binding protein, Zm1a-6 RNA | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3ORG
 
 | |
3U7R
 
 | | FerB - flavoenzyme NAD(P)H:(acceptor) oxidoreductase (FerB) from Paracoccus denitrificans | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NADPH-dependent FMN reductase, NONAETHYLENE GLYCOL | | Authors: | Marek, J, Klumpler, T, Sedlacek, V, Kucera, I. | | Deposit date: | 2011-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structural and Functional Basis of Catalysis Mediated by NAD(P)H:acceptor Oxidoreductase (FerB) of Paracoccus denitrificans.
Plos One, 9, 2014
|
|
4N2Q
 
 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JVP
 
 | |
3OLZ
 
 | |
6JB6
 
 | | Crystal structure of Ub-conjugated Ube2K C92K&K97A mutant (isopeptide linkage), 2.7 A resolution | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Lee, J.-G, Youn, H.-S, Lee, Y, An, J.Y, Park, K.R, Kang, J.Y, Mun, S.A, Park, J, Park, T, Jin, M.W, Yang, J, Eom, S.H. | | Deposit date: | 2019-01-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Ub-conjugated Ube2K C92K&K97A mutant (isopeptide linkage), 2.7 A resolution
To Be Published
|
|
1QHX
 
 | |
4JH7
 
 | | Crystal Structure of FosB from Bacillus cereus with Manganese and L-Cysteine-Fosfomycin Product | | Descriptor: | (2R)-2-azanyl-3-[(1R,2S)-2-oxidanyl-1-phosphono-propyl]sulfanyl-propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
8PW6
 
 | | C respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
