2VAB
 
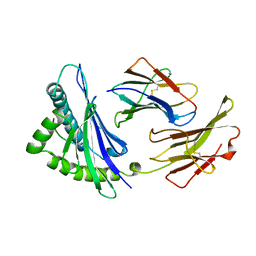 | |
8BP8
 
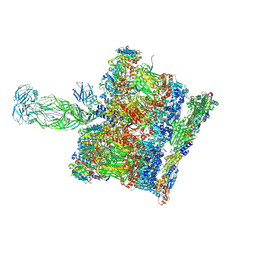 | | SPA of Trypsin untreated Rotavirus TLP spike | | Descriptor: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
3K8E
 
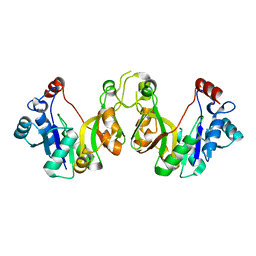 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
2VAA
 
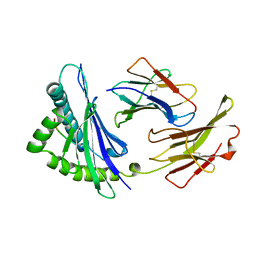 | |
7WHW
 
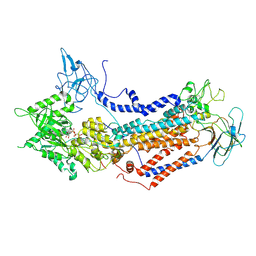 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
4Z7W
 
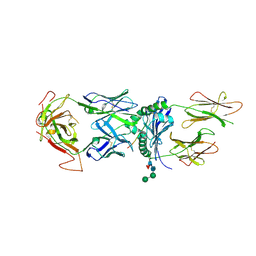 | | T316 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQ8-glia-alpha1, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
7WHV
 
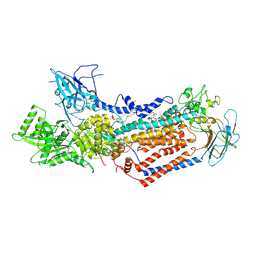 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
3J24
 
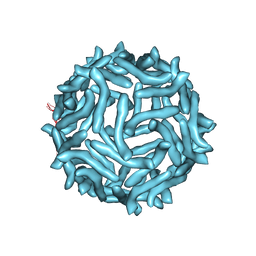 | | CryoEM reconstruction of complement decay-accelerating factor | | Descriptor: | Complement decay-accelerating factor | | Authors: | Yoder, J.D, Hafenstein, S.H. | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
7ADP
 
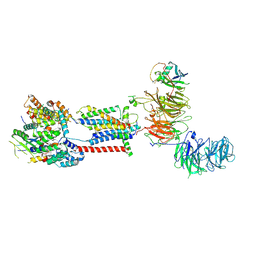 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
1O3S
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
6ZLW
 
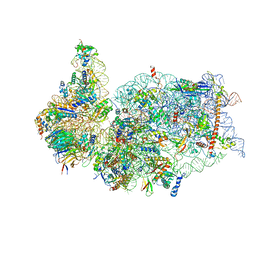 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
1O64
 
 | |
6ZP1
 
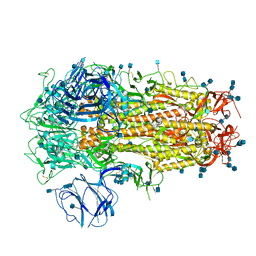 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8AKN
 
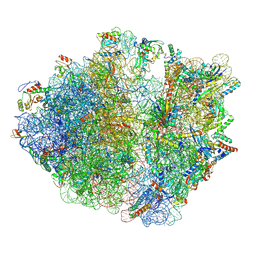 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8AFP
 
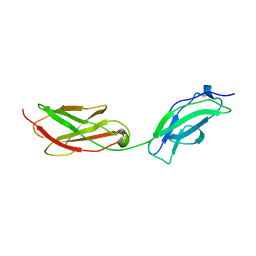 | | Structure of fibronectin 2 and 3 of L1CAM at 3.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neural cell adhesion molecule L1 | | Authors: | Guedez, G, Loew, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1.
Faseb J., 37, 2023
|
|
8ANA
 
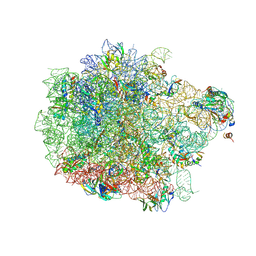 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the 50S ribosomal subunit | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
1O62
 
 | |
8AM9
 
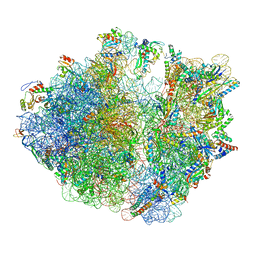 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the elongating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
6Y6X
 
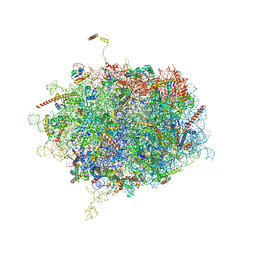 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
4ZG6
 
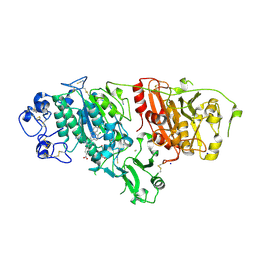 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4WVE
 
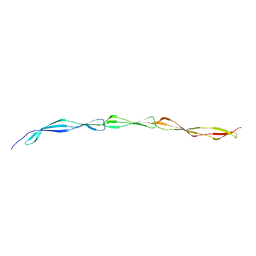 | |
4WWP
 
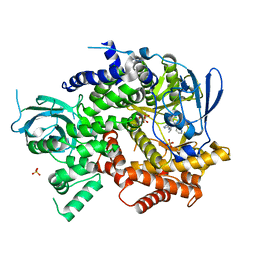 | | Crystal structure of human PI3K-gamma in complex with pyridinylquinoline inhibitor N-{(1S)-1-[8-chloro-2-(2-methylpyridin-3-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine | | Descriptor: | GLYCEROL, N-{(1S)-1-[8-chloro-2-(2-methylpyridin-3-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, ... | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
7TNT
 
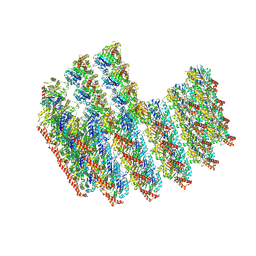 | | The tubulin-based conoid from detergent-extract Toxoplasma gondii cells | | Descriptor: | Tubulin alpha chain, Tubulin beta chain | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TNS
 
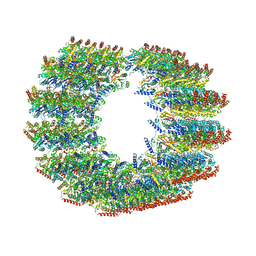 | | Subpellicular microtubule from detergent-extract Toxoplasma gondii cells | | Descriptor: | Microtubule associated protein SPM1, PDI family protein, Tubulin alpha chain, ... | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5U9Z
 
 | |
