3D7J
 
 | | SCO6650, a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein SCO6650 | | Authors: | Spoonamore, J.E, Roberts, S.A, Heroux, A, Bandarian, V. | | Deposit date: | 2008-05-21 | | Release date: | 2008-10-21 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4FWZ
 
 | | Aquoferric CuB myoglobin (L29H F43H sperm whale myoglobin) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gao, Y.-G, Robinson, H, Petrik, I.D, Miner, K.D, Lu, Y. | | Deposit date: | 2012-07-02 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Designed Functional Metalloenzyme that Reduces O(2) to H(2) O with Over One Thousand Turnovers.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3D3N
 
 | | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Putative lipase/esterase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Ciccosanti, C, Maglaqui, M, Foote, E.L, Zhao, L, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108
To be Published
|
|
3LSF
 
 | | Piracetam bound to the ligand binding domain of GluA2 | | Descriptor: | 2-(2-oxopyrrolidin-1-yl)acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
3LT8
 
 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding, crystallized in the presence of 100 mM ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LV2
 
 | |
3LFI
 
 | |
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
3CEY
 
 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
4PKI
 
 | |
5VV6
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-(((4-(Dimethylamino)phenethyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[({2-[4-(dimethylamino)phenyl]ethyl}amino)methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VWY
 
 | |
5VVH
 
 | | Crystal Structure of the Effector Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens | | Descriptor: | FORMIC ACID, Octopine catabolism/uptake operon regulatory protein OccR, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
3COL
 
 | |
5VX3
 
 | | Bcl-xL in complex with Bim-h3Pc-RT | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein 1, Bcl-2-like protein 11 | | Authors: | Cowan, A.D, Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
6LF2
 
 | | SeviL bound to asialo-GM1 saccharide | | Descriptor: | SeviL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
3LT9
 
 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
4PSC
 
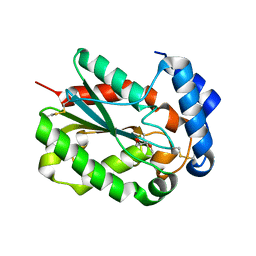 | | Structure of cutinase from Trichoderma reesei in its native form. | | Descriptor: | Carbohydrate esterase family 5, GLYCEROL | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
3CRN
 
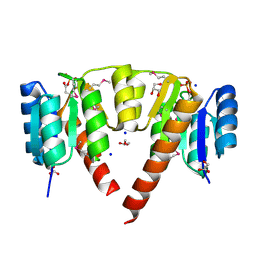 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, Response regulator receiver domain protein, CheY-like, ... | | Authors: | Hiner, R.L, Toro, R, Patskovsky, Y, Freeman, J, Chang, S, Smith, D, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1.
To be Published
|
|
4FWY
 
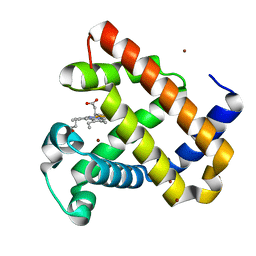 | | F33Y CuB myoglobin (F33Y L29H F43H sperm whale myoglobin) with copper bound | | Descriptor: | COPPER (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gao, Y.-G, Robinson, H, Petrik, I.D, Miner, K.D, Lu, Y. | | Deposit date: | 2012-07-02 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Designed Functional Metalloenzyme that Reduces O(2) to H(2) O with Over One Thousand Turnovers.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GDX
 
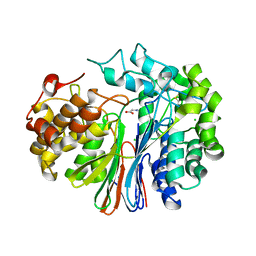 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
3CPG
 
 | |
4P38
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with AZD8329 | | Descriptor: | 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Ogg, D, Hargreaves, D, Gerhardt, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel acidic 11 beta-hydroxysteroid dehydrogenase type 1 (11 beta-HSD1) inhibitor with reduced acyl glucuronide liability: the discovery of 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid (AZD8329).
J.Med.Chem., 55, 2012
|
|
3CSL
 
 | |
3LSL
 
 | | Piracetam bound to the ligand binding domain of GluA2 (flop form) | | Descriptor: | 2-(2-oxopyrrolidin-1-yl)acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.122 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
