8H6J
 
 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6E
 
 | | Cryo-EM structure of human exon-defined spliceosome in the late pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6L
 
 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
8COB
 
 | |
9LDT
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9FQ0
 
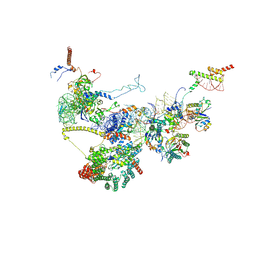 | | Human NatA-NAC-MAP1 80S ribosome complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Multi-protein assemblies orchestrate enzymatic processing of the nascent chain on the 80S ribosome
To Be Published
|
|
9FA4
 
 | |
9LDB
 
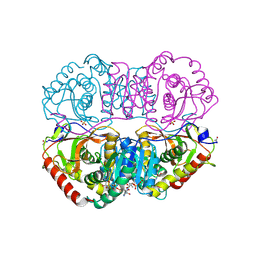 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9FA7
 
 | |
9FBY
 
 | |
9FPZ
 
 | | Human NatA-MAP2 80S ribosome complex | | Descriptor: | 28S rRNA (300-MER, fragments), 5.8S rRNA (58-MER), ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multi-protein assemblies orchestrate enzymatic processing of the nascent chain on the 80S ribosome
To Be Published
|
|
8CKF
 
 | | Crystal Structure of the first bromodomain of human BRD4 L94C variant in complex with racemic 3,5-dimethylisoxazol ligand | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{S})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Thomas, A.M, McDonough, M.A, Schiedel, M, Conway, S.J. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutate and Conjugate: A Method to Enable Rapid In-Cell Target Validation.
Acs Chem.Biol., 18, 2023
|
|
8UPF
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8WAK
 
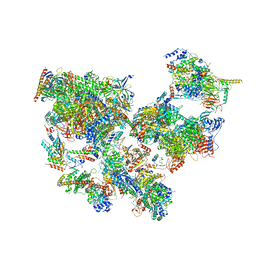 | | Structure of transcribing complex 2 (TC2), the initially transcribing complex with Pol II positioned 2nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAR
 
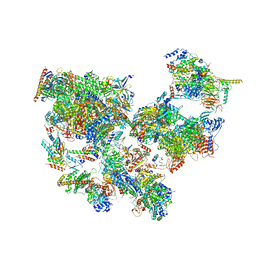 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8W9E
 
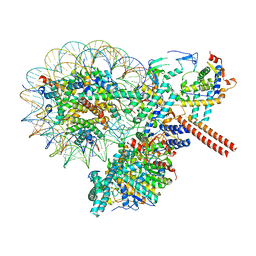 | |
8W9D
 
 | |
8X73
 
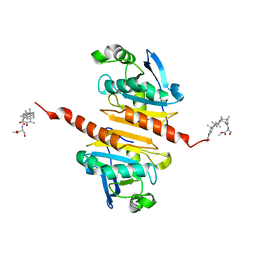 | | Crystal structure of Peroxiredoxin I in complex with compound 19-069 | | Descriptor: | Peroxiredoxin-1, methyl (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]-3-oxidanyl-propanoate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X71
 
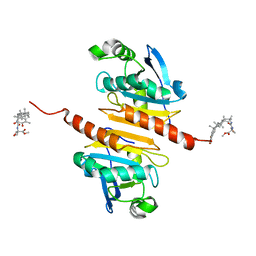 | | Crystal structure of Peroxiredoxin I in complex with compound 19-064 | | Descriptor: | Peroxiredoxin-1, methyl 3-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]oxetane-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8W50
 
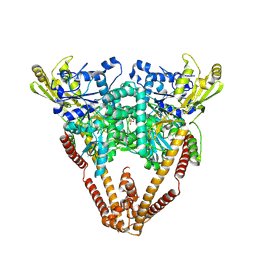 | |
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8W8F
 
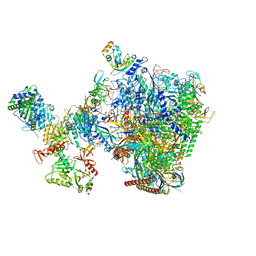 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8UIS
 
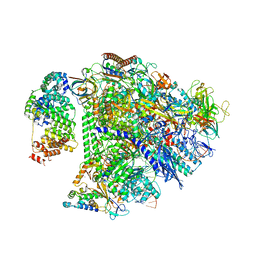 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
