7Q6X
 
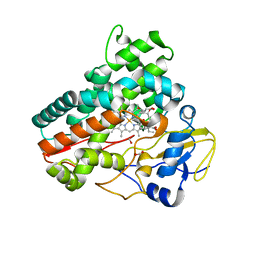 | | OleP mutant S240Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q89
 
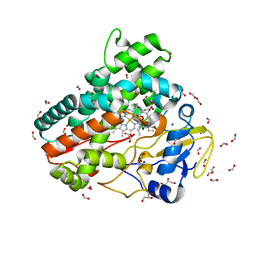 | | OleP mutant G92W in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q6R
 
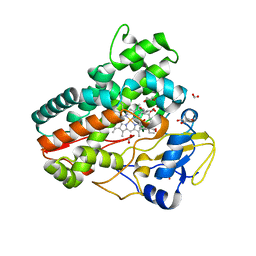 | | OleP mutant E89Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
6MHN
 
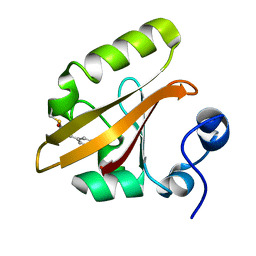 | | Photoactive Yellow Protein with covalently bound 3-chloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3-chloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
7SO5
 
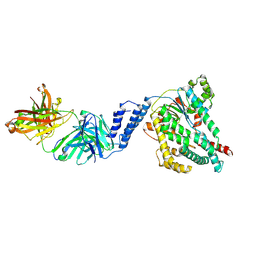 | |
8Q34
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the ligand ZZ001229a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1~{H}-imidazo[4,5-b]pyridin-2-ylmethyl)-3-(3-methyl-1,2-diazirin-3-yl)propanamide | | Authors: | MacLean, E.M, Gao, Q, Williams, E, Balcomb, B.H, von Delft, F, Bajusz, D, Keeley, A, Abranyi-Balogh, P, Koekemoer, L, Keseru, G.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein binding sites by photoreactive fragment pharmacophores.
Commun Chem, 7, 2024
|
|
6MHI
 
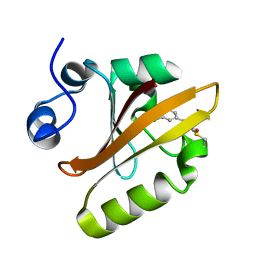 | | Photoactive Yellow Protein with covalently bound 3,5-dichloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3,5-dichloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
8CAI
 
 | | Streptomycin and Hygromycin B bound to the 30S body | | Descriptor: | 16S rRNA, 30S ribosomal protein S16, 30S ribosomal protein S2, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
8ESY
 
 | | D30N mutant HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
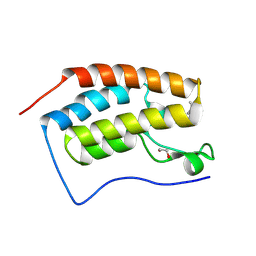
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
8ELM
 
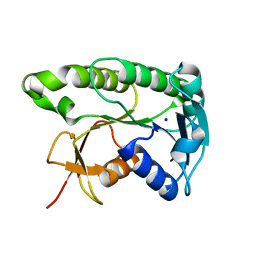 | | Apo human biliverdin reductase beta (293K) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
6Y8N
 
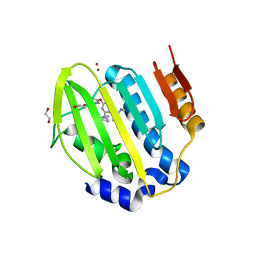 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
8ELL
 
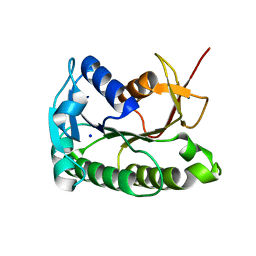 | | Apo human biliverdin reductase beta (cryogenic) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
8QEO
 
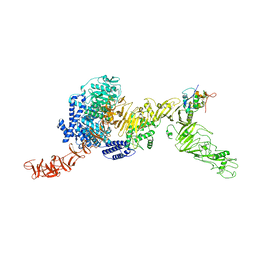 | |
8QEN
 
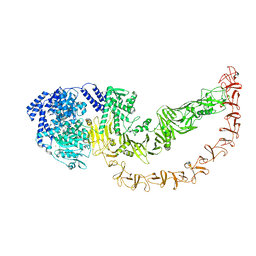 | |
9FT8
 
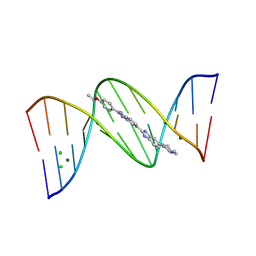 | | Crystal structure of d(CGTGAATTCACG) with HT1 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*GP*AP*AP*TP*TP*CP*AP*CP*G)-3'), ... | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L. | | Deposit date: | 2024-06-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of B-DNA d(CGTGAATTCACG)2 in Complex with the Specific Minor Groove Binding Fluorescent Marker Hoechst 33342
Crystals, 15, 2025
|
|
6MKT
 
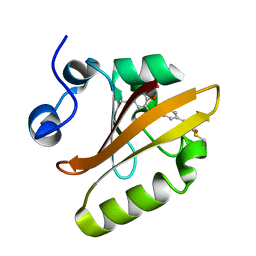 | | Photoactive Yellow Protein with 3-chlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
8TEA
 
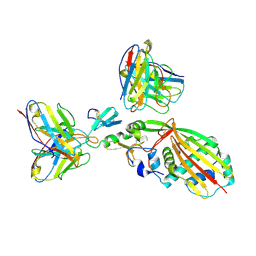 | |
8EE1
 
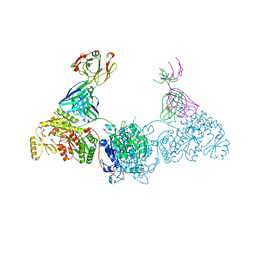 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment AA5 | | Descriptor: | 6-deoxyerythronolide B synthase, AA5 antibody heavy chain, AA5 antibody light chain | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
9EUO
 
 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 168, 2024
|
|
8P00
 
 | | Cryo-EM structure of Rotavirus B NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Chamera, S, Nowotny, M. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of rotavirus B NSP2 reveals its unique tertiary architecture.
J.Virol., 98, 2024
|
|
5E5Z
 
 | |
6Z3M
 
 | | Repulsive Guidance Molecule B (RGMB) in complex with Growth Differentiation Factor 5 (GDF5) and Neogenin 1 (NEO1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 5, Neogenin, ... | | Authors: | Malinauskas, T, Peer, T.V, Bishop, B, Muller, T.D, Siebold, C. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (5.501 Å) | | Cite: | Repulsive guidance molecules lock growth differentiation factor 5 in an inhibitory complex.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QFQ
 
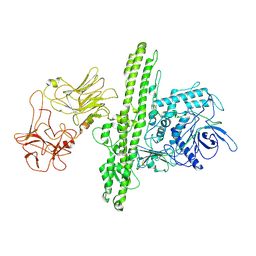 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
