6FXA
 
 | |
5I88
 
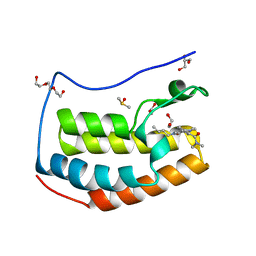 | | BRD4 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 1,2-ETHANEDIOL, 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
7TIN
 
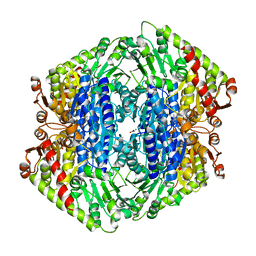 | | The Structure of S. aureus MenD | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Stanborough, T, Ho, N.A.T, Akazong, E.W, Jiao, W. | | Deposit date: | 2022-01-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus MenD by 1,4-dihydroxy naphthoic acid: a feedback inhibition mechanism of the menaquinone biosynthesis pathway.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 378, 2023
|
|
5KJ8
 
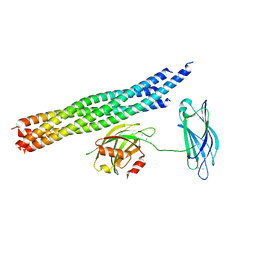 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from synchrotron diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
5KMM
 
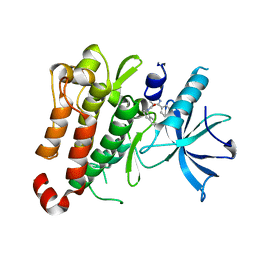 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(1-naphthyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-naphthalen-1-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9H8F
 
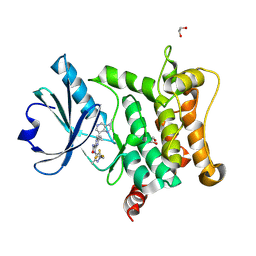 | | Crystal structure of HPK1 T165E/S171E in complex with pyrazine carboxamide inhibitor AZ3246 (compound 24) | | Descriptor: | 1,2-ETHANEDIOL, 5-cyclopropyl-6-(3-methylimidazo[4,5-c]pyridin-7-yl)-3-[[3-methyl-1-[2,2,2-tris(fluoranyl)ethyl]pyrazol-4-yl]amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M, Pflug, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
6G40
 
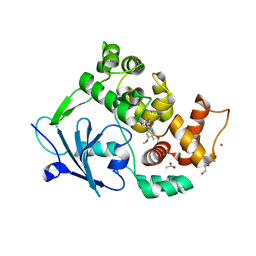 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
3M67
 
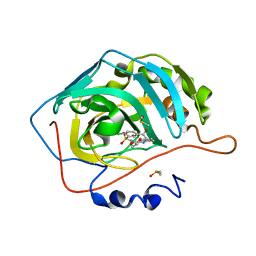 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
5BTN
 
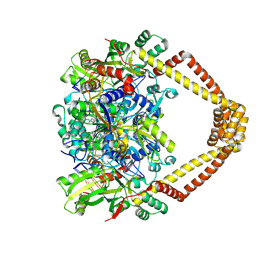 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methyl-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B05
 
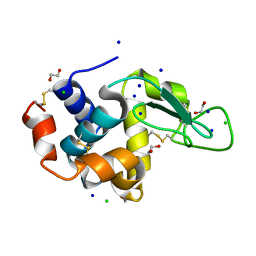 | | Lysozyme (control experiment) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5BTA
 
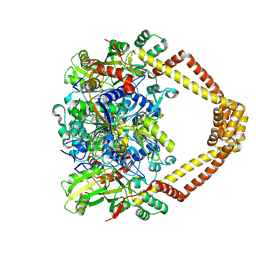 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZZ6
 
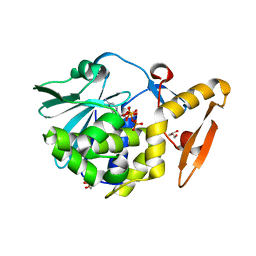 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine triphosphate at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5VI9
 
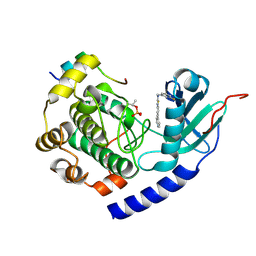 | |
5LQD
 
 | | Trehalose-6-phosphate synthase, GDP-glucose-dependent OtsA | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase | | Authors: | Miah, F, Asencion Diez, M.D, Stevenson, C.E.M, Lawson, D.M, Iglesias, A.A, Bornemann, S. | | Deposit date: | 2016-08-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Production and Utilization of GDP-glucose in the Biosynthesis of Trehalose 6-Phosphate by Streptomyces venezuelae.
J. Biol. Chem., 292, 2017
|
|
7ZCZ
 
 | |
5ZKA
 
 | | Crystal structure of N-acetylneuraminate lyase from Fusobacterium nucleatum complexed with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, N-acetylneuraminate lyase, TRIETHYLENE GLYCOL | | Authors: | Kumar, J.P, Rao, H, Nayak, V, Ramaswamy, S. | | Deposit date: | 2018-03-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7Z1U
 
 | | Biochemical implications of the substitution of a unique cysteine residue in sugar beet phytoglobin BvPgb 1.2 | | Descriptor: | Non-symbiotic hemoglobin class 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nyblom, M, Christensen, S, Leiva Eriksson, N, Bulow, L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
1KD0
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
5CTB
 
 | |
7ZER
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
6C4P
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PMP Complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Berkowitz, D.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
5I5X
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.65A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIAZOLE COMPOUND AND PMP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 5-methyl-4-oxo-N-(1,3,4-thiadiazol-2-yl)-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxamide, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5BOP
 
 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5BTL
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methyl-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3MWH
 
 | | The 1.4 Ang crystal structure of the ArsD arsenic metallochaperone provides insights into its interactions with the ArsA ATPase | | Descriptor: | Arsenical resistance operon trans-acting repressor arsD, GLYCEROL | | Authors: | Ye, J, Ajees, A.A, Yang, J, Rosen, B.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase.
Biochemistry, 49, 2010
|
|
