3ZQM
 
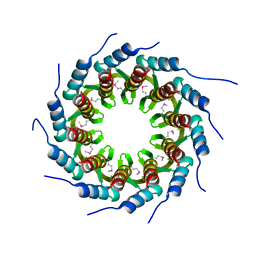 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1GFF
 
 | |
7KIX
 
 | |
6NTU
 
 | |
1QRI
 
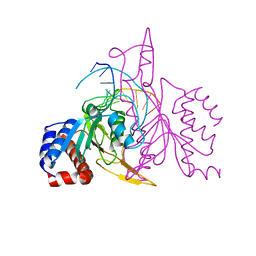 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN E144D MUTATION AT 2.7 A | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
5DD8
 
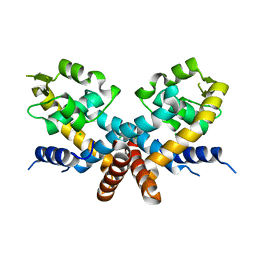 | | The Crystal structure of HucR mutant (HucR-E48Q) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family | | Authors: | Deochand, D.K, Perera, I.C, Crochet, R.B, Gilbert, N.C, Newcomer, M.E, Grove, A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices.
Mol Biosyst, 12, 2016
|
|
4NBP
 
 | |
4DOV
 
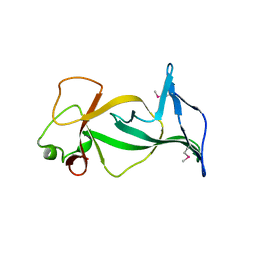 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
1SAP
 
 | |
1QRH
 
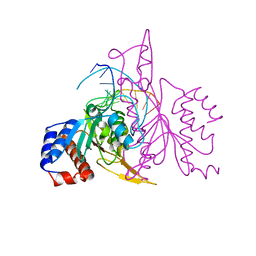 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN R145K MUTATION AT 2.7 A | | Descriptor: | 5'-(TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1F1Z
 
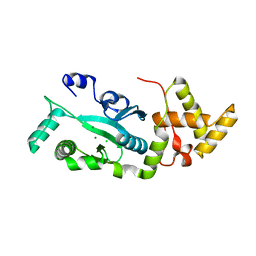 | | TNSA, a catalytic component of the TN7 transposition system | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TNSA ENDONUCLEASE | | Authors: | Hickman, A.B, Li, Y, Mathew, S.V, May, E.W, Craig, N.L, Dyda, F. | | Deposit date: | 2000-05-21 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unexpected structural diversity in DNA recombination: the restriction endonuclease connection.
Mol.Cell, 5, 2000
|
|
1Z91
 
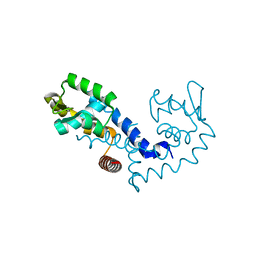 | |
2HAX
 
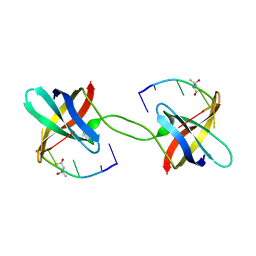 | |
1IH2
 
 | |
1IH3
 
 | |
6EKO
 
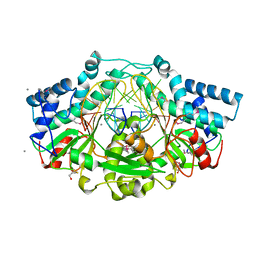 | | Crystal structure of Type IIP restriction endonuclease PfoI with cognate DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*CP*CP*CP*GP*GP*AP*GP*CP*GP*T)-3'), Restriction endonuclease PfoI | | Authors: | Tamulaitiene, G, Manakova, E, Jovaisaite, V, Grazulis, S, Siksnys, V. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Unique mechanism of target recognition by PfoI restriction endonuclease of the CCGG-family.
Nucleic Acids Res., 47, 2019
|
|
3W0F
 
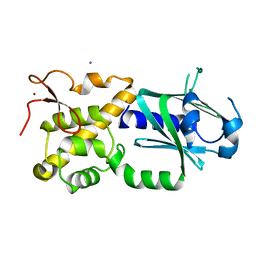 | | Crystal structure of mouse Endonuclease VIII-LIKE 3 (mNEIL3) | | Descriptor: | Endonuclease 8-like 3, IODIDE ION, ZINC ION | | Authors: | Liu, M, Imamura, K, Averill, A.M, Wallace, S.S, Doublie, S. | | Deposit date: | 2012-10-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Mouse Ortholog of Human NEIL3 with a Marked Preference for Single-Stranded DNA
Structure, 21, 2013
|
|
9EOX
 
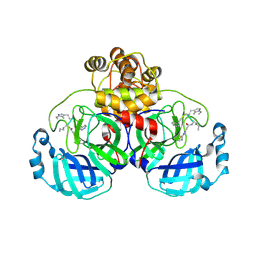 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | (2~{S})-2-[[(3~{R})-3-[[(2~{R})-2-[(2-cyanopyridin-4-yl)carbonylamino]hexanoyl]amino]-4-(1~{H}-indol-3-yl)butanoyl]amino]-3-pyridin-4-yl-propanoic acid, 3C-like proteinase nsp5, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
328D
 
 | | STRUCTURE OF A D(CGCGAATTCGCG)2-SN7167 COMPLEX | | Descriptor: | 4-[4-[2-AMINO-4-[4,6-(N-METHYLQUINOLINIUM)AMINO]BENZAMIDO]ANILINO]-N-METHYLPYRIDINIUM MESYLATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Clark, G.R, Denny, W.A. | | Deposit date: | 1997-04-15 | | Release date: | 1997-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Minor groove binding of a bis-quaternary ammonium compound: the crystal structure of SN 7167 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 25, 1997
|
|
4LMD
 
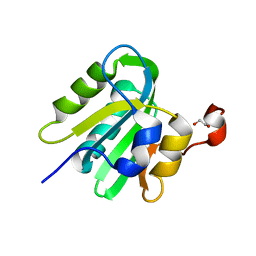 | |
2HIN
 
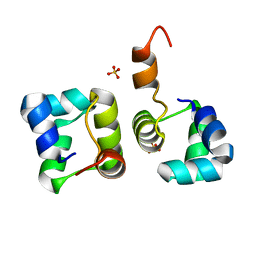 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
7WE6
 
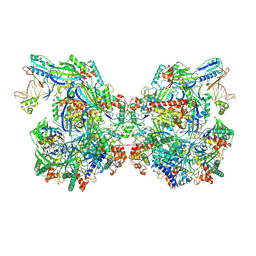 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
9EO6
 
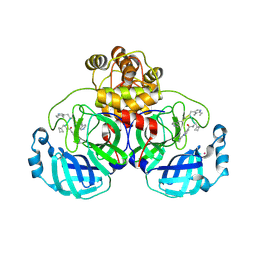 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, ~{N}-[(2~{S})-3-cyclobutyl-1-[[(2~{R})-1-(1~{H}-indol-3-yl)-4-[[(2~{R})-1-(methylamino)-1-oxidanylidene-3-pyridin-4-yl-propan-2-yl]amino]-4-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-(iminomethyl)pyridine-4-carboxamide | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 2-cyano-~{N}-[(2~{R})-1-[[(2~{S})-1-(1~{H}-indol-3-yl)-4-[[(2~{R})-1-(methylamino)-1-oxidanylidene-3-pyridin-4-yl-propan-2-yl]amino]-4-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-hexan-2-yl]pyridine-4-carboxamide, 3C-like proteinase nsp5, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
