2PWY
 
 | | Crystal Structure of a m1A58 tRNA methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (adenine-N(1)-)-methyltransferase | | Authors: | Barraud, P, Golinelli-Pimpaneau, B, Tisne, C. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermus thermophilus tRNA m(1)A(58) Methyltransferase and Biophysical Characterization of Its Interaction with tRNA.
J.Mol.Biol., 377, 2008
|
|
4ZJ2
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli :E166N mutant | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, C.S, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZQS
 
 | | New compact conformation of linear Ub2 structure | | Descriptor: | ubiquitin | | Authors: | Thach, T.T, Shin, D, Han, S, Lee, S. | | Deposit date: | 2015-05-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3BZJ
 
 | | UVDE K229L | | Descriptor: | MANGANESE (II) ION, SULFATE ION, UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
3C0Q
 
 | | UVDE E175A | | Descriptor: | MANGANESE (II) ION, SULFATE ION, UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
1URW
 
 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4OH5
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OHA
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4ZJ3
 
 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OGH
 
 | | Crystal structure of T877A-AR-LBD | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, SULFATE ION | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2Q6H
 
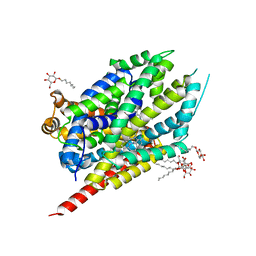 | | Crystal Structure Analysis of LeuT complexed with L-leucine, sodium, and clomipramine | | Descriptor: | 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, LEUCINE, SODIUM ION, ... | | Authors: | Singh, S.K, Yamashita, A, Gouaux, E. | | Deposit date: | 2007-06-05 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antidepressant binding site in a bacterial homologue of neurotransmitter transporters.
Nature, 448, 2007
|
|
1U71
 
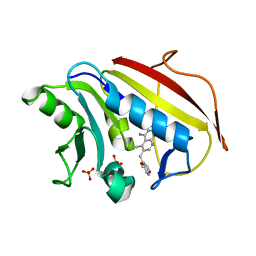 | | Understanding the Role of Leu22 Variants in Methotrexate Resistance: Comparison of Wild-type and Leu22Arg Variant Mouse and Human Dihydrofolate Reductase Ternary Crystal Complexes with Methotrexate and NADPH | | Descriptor: | 6-(2,5-DIMETHOXY-BENZYL)-5-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2004-08-02 | | Release date: | 2005-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2XR7
 
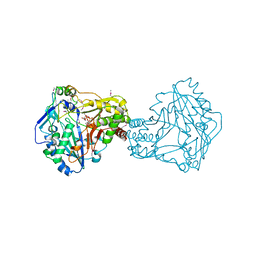 | | Crystal Structure of Nicotiana tabacum malonyltransferase (NtMat1) complexed with malonyl-coa | | Descriptor: | MALONYL-COENZYME A, MALONYLTRANSFERASE | | Authors: | Manjasetty, B.A, Yu, X.H, Panjikar, S, Taguchi, G, Chance, M.R, Liu, C.J. | | Deposit date: | 2010-09-11 | | Release date: | 2011-09-21 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Modification of Flavonol and Naphthol Glucoconjugates by Nicotiana Tabacum Malonyltransferase (Ntmat1).
Planta, 236, 2012
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
3AKA
 
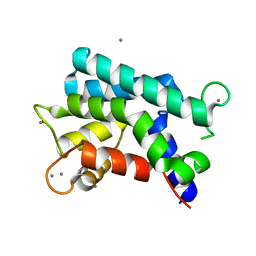 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
2V69
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit mutation D473E | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2-O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
6I45
 
 | | Crystal structure of I13V/I62V/V77I South African HIV-1 subtype C protease containing a D25A mutation | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, SODIUM ION | | Authors: | Sherry, D, Pandian, R, Achilonu, I.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-active site mutations in the HIV protease: Diminished drug binding affinity is achieved through modulating the hydrophobic sliding mechanism.
Int.J.Biol.Macromol., 217, 2022
|
|
6I4A
 
 | | Structure of P. aeruginosa LpxC with compound 18d: (2R)-N-Hydroxy-4-(6-((1-(hydroxymethyl)cyclopropyl)buta-1,3-diyn-1-yl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-[4-[1-(hydroxymethyl)cyclopropyl]buta-1,3-diynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
4N71
 
 | | X-Ray Crystal Structure of 2-amino-1-hydroxyethylphosphonate-bound PhnZ | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | Woersdoerfer, B, Lingaraju, M, Yennawar, N, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.-E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
7Z0Y
 
 | | THSC20.HVTR04 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
6I48
 
 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
