6O5J
 
 | | Crystal Structure of DAD2 bound to quinazolinone derivative | | Descriptor: | 1-(4-hydroxy-3-nitrophenyl)quinazoline-2,4(1H,3H)-dione, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamiaux, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode.
Biochem.J., 476, 2019
|
|
6OL9
 
 | | Structure of the M5 muscarinic acetylcholine receptor (M5-T4L) bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Vuckovic, Z, Christopoulos, A, Thal, D.M. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Crystal structure of the M5muscarinic acetylcholine receptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OLG
 
 | |
6OY9
 
 | | Structure of the Rhodopsin-Transducin Complex | | Descriptor: | Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
6ON6
 
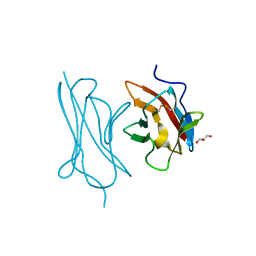 | | Crystal Structure of the RIG-5 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NeuRonal IgCAM-5 | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ONF
 
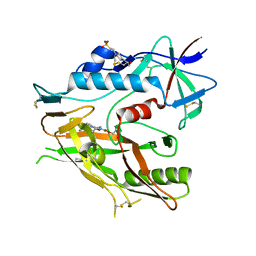 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A02. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6OR9
 
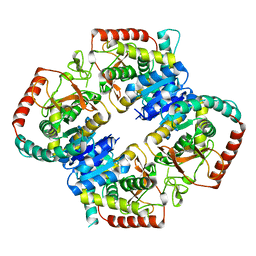 | |
8V1T
 
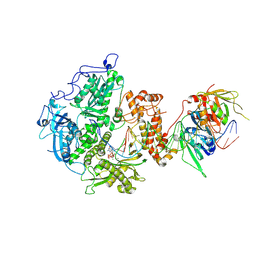 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
6OXN
 
 | |
8V1R
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
6OYU
 
 | | Structure of an ancestral-reconstructed cytochrome P450 1B1 with alpha-naphthoflavone | | Descriptor: | 2-PHENYL-4H-BENZO[H]CHROMEN-4-ONE, Cytochrome P450 1B1, GLYCEROL, ... | | Authors: | Bart, A.G, Harris, K.L, Scott, E.E. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of an ancestral mammalian family 1B1 cytochrome P450 with increased thermostability.
J.Biol.Chem., 295, 2020
|
|
5UE5
 
 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
8V1Q
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1S
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
5UE2
 
 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
8V23
 
 | |
8V5I
 
 | | Crystal structure of MAP4K4 in complex with an inhibitor | | Descriptor: | (3M)-N~6~-(1,4-dimethyl-1H-pyrazol-3-yl)-3-(1-methyl-1H-imidazol-5-yl)-2,7-naphthyridine-1,6-diamine, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Greasley, S.E, Diehl, W. | | Deposit date: | 2023-11-30 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Microtubule-Associated Serine/Threonine Kinase-like (MASTL).
J.Med.Chem., 67, 2024
|
|
5JPW
 
 | | Molecular basis for protein recognition specificity of the DYNLT1/Tctex1 canonical binding groove. Characterization of the interaction with activin receptor IIB | | Descriptor: | Dynein light chain Tctex-type 1,Cytoplasmic dynein 1 intermediate chain 2 | | Authors: | Rodriguez-Crespo, I, Merino-Gracia, J, Bruix, M, Zamora-Carreras, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for the Protein Recognition Specificity of the Dynein Light Chain DYNLT1/Tctex1: CHARACTERIZATION OF THE INTERACTION WITH ACTIVIN RECEPTOR IIB.
J.Biol.Chem., 291, 2016
|
|
5W0B
 
 | | Structure of human TUT7 catalytic module (CM) | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7RMJ
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor (S)-N-(1-(3-(4-chloro-3-(methylsulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl)-6-(3-methyl-3-(methylsulfonyl)but-1-yn-1-yl)pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CAPSID PROTEIN P24, CHLORIDE ION, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Mechanistic Bases of Viral Resistance to HIV-1 Capsid Inhibitor Lenacapavir.
Mbio, 13, 2022
|
|
5VX0
 
 | | Bak in complex with Bim-h3Glg | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 11, ... | | Authors: | Brouwer, J.M, Lan, P, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
7RMM
 
 | |
5VTP
 
 | |
5VQ9
 
 | | Structure of human TRIP13, Apo form | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5VWW
 
 | | Bak core latch dimer in complex with Bim-RT - Tetragonal | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BROMIDE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
