2OMQ
 
 | |
4FLC
 
 | | Structural and Biochemical Characterization of Human Adenylosuccinate Lyase (ADSL) and the R303C ADSL Deficiency Associated Mutation | | Descriptor: | Adenylosuccinate lyase | | Authors: | Deaton, M.K, Ray, S.P, Capodagli, G.C, Calkins, L.A.F, Sawle, L, Ghosh, K, Patterson, D, Pegan, S.D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Characterization of Human Adenylosuccinate Lyase (ADSL) and the R303C ADSL Deficiency-Associated Mutation.
Biochemistry, 51, 2012
|
|
2OMY
 
 | | Crystal structure of InlA S192N/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
4FRR
 
 | |
2ON6
 
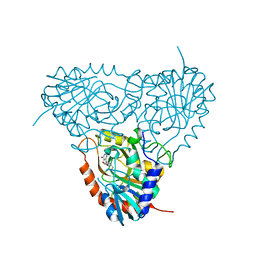 | | Crystal structure of human purine nucleoside phosphorylase mutant H257F with Imm-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Murkin, A.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Neighboring Group Participation in the Transition State of Human Purine Nucleoside Phosphorylase
Biochemistry, 46, 2007
|
|
4FRI
 
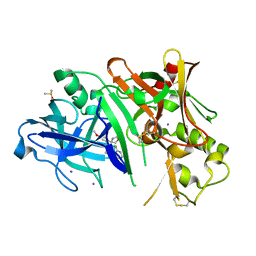 | | Crystal structure of BACE1 in complex with biarylspiro aminooxazoline 6 | | Descriptor: | (4R)-4-[3-(2-fluoropyridin-3-yl)phenyl]-4-(4-methoxyphenyl)-4,5-dihydro-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure- and Property-Based Design of Aminooxazoline Xanthenes as Selective, Orally Efficacious, and CNS Penetrable BACE Inhibitors for the Treatment of Alzheimer's Disease.
J.Med.Chem., 55, 2012
|
|
2ONC
 
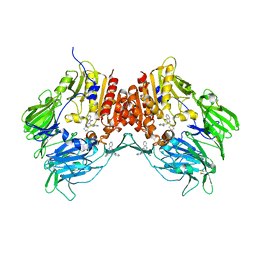 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
3QLS
 
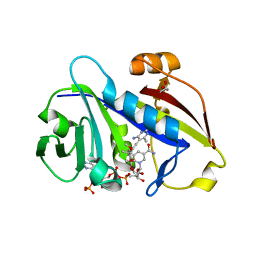 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
2OLP
 
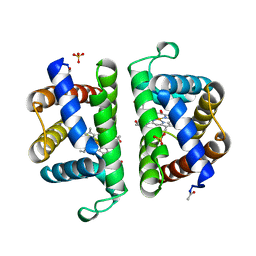 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2007-01-19 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
2OMU
 
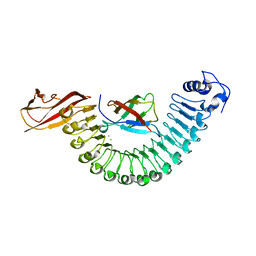 | | Crystal structure of InlA G194S+S Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4FHN
 
 | |
2OOV
 
 | |
3QQ7
 
 | |
3QTL
 
 | |
3QQM
 
 | |
2OMW
 
 | | Crystal structure of InlA S192N Y369S/mEC1 complex | | Descriptor: | CHLORIDE ION, Epithelial-cadherin, Internalin-A | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
4F62
 
 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2FS2
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
3QUE
 
 | | Human p38 MAP Kinase in Complex with Skepinone-L | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Mayer-Wrangowski, S, Richters, A, Rauh, D. | | Deposit date: | 2011-02-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor.
Nat.Chem.Biol., 8, 2012
|
|
3QTI
 
 | | c-Met Kinase in Complex with NVP-BVU972 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-b]pyridazin-3-yl]methyl}quinoline, CHLORIDE ION, GLYCEROL, ... | | Authors: | Appleton, B.A. | | Deposit date: | 2011-02-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Drug Resistance Screen Using a Selective MET Inhibitor Reveals a Spectrum of Mutations That Partially Overlap with Activating Mutations Found in Cancer Patients.
Cancer Res., 71, 2011
|
|
2OO4
 
 | | Structure of LNR-HD (Negative Regulatory Region) from human Notch 2 | | Descriptor: | CALCIUM ION, GLYCEROL, Neurogenic locus notch homolog protein 2, ... | | Authors: | Gordon, W.R, Vardar-Ulu, D, Histen, G, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for autoinhibition of Notch
Nat.Struct.Mol.Biol., 14, 2007
|
|
2FS8
 
 | | Human beta-tryptase II with inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
4F75
 
 | |
4FOO
 
 | |
3QC0
 
 | |
