2KUY
 
 | | Structure of Glycocin F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prebacteriocin glycocin F | | Authors: | Venugopal, H, Edwards, P, Schwalbe, M, Claridge, J, Stepper, J, Patchett, M, Loo, T, Libich, D, Norris, G, Pascal, S. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamic, and chemical characterization of a novel s-glycosylated bacteriocin.
Biochemistry, 50, 2011
|
|
6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6F0Q
 
 | |
6F0Y
 
 | | Rtt109 peptide bound to Asf1 | | Descriptor: | Histone chaperone ASF1, histone acetyltransferase Rtt109 C-terminus | | Authors: | Lercher, L, Kirkpatrick, J.P, Carlomagno, T. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the Asf1-Rtt109 interaction and its role in histone acetylation.
Nucleic Acids Res., 46, 2018
|
|
3KHG
 
 | | Dpo4 extension ternary complex with misinserted A opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-AMINOFLUORENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6PJB
 
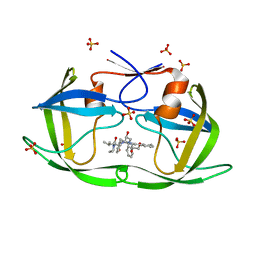 | | HIV-1 Protease NL4-3 WT in Complex with Lopinavir | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJN
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6F43
 
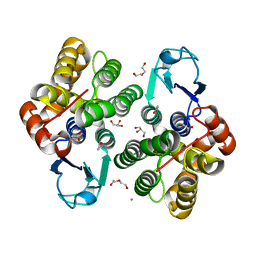 | | Crystal structure of apo form of Glutathione transferase Omega 3S from Trametes versicolor | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
7RTP
 
 | |
4XPZ
 
 | |
6F4G
 
 | | 'Crystal structure of the Drosophila melanogaster SNF/U2A'/U2-SL4 complex | | Descriptor: | CHLORIDE ION, Probable U2 small nuclear ribonucleoprotein A', SULFATE ION, ... | | Authors: | Weber, G, DeKoster, G, Holton, N, Hall, K.B, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6F2R
 
 | | A heterotetramer of human HspB2 and HspB3 | | Descriptor: | Heat shock protein beta-2, Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-3,Heat shock protein beta-2, HspB2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2,Heat shock protein beta-2, ... | | Authors: | Clark, A.R, Cole, A.R, Boelens, W.C, Keep, N.H, Slingsby, C. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Terminal Regions Confer Plasticity to the Tetrameric Assembly of Human HspB2 and HspB3.
J.Mol.Biol., 430, 2018
|
|
6F55
 
 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
6F5O
 
 | |
6P24
 
 | | Escherichia coli tRNA synthetase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
6P2R
 
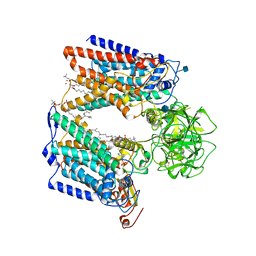 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PBI
 
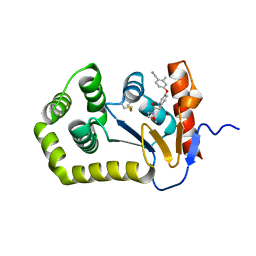 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6P3P
 
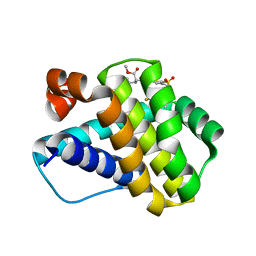 | | Crystal structure of Mcl-1 in complex with compound 65 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, methyl N-(5-{[2-chloro-5-(trifluoromethyl)phenyl]sulfamoyl}-4-methylthiophene-2-carbonyl)-D-phenylalaninate | | Authors: | Toms, A.V, Follows, B. | | Deposit date: | 2019-05-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of novel biaryl sulfonamide based Mcl-1 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6P3Q
 
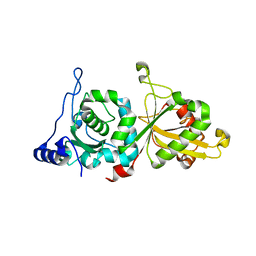 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
4XTU
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
4XU2
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 87 with a 3'deoxy ribose | | Descriptor: | 3',5'-dideoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8500284 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XUF
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor quizartinib (AC220) | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zorn, J.A, Wang, Q, Fujimura, E, Barros, T, Kuriyan, J. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the FLT3 Kinase Domain Bound to the Inhibitor Quizartinib (AC220).
Plos One, 10, 2015
|
|
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
6P5D
 
 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6FAO
 
 | | Discovery and characterization of a thermostable GH6 endoglucanase from a compost metagenome | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 6, SULFATE ION | | Authors: | Jensen, M.S, Fredriksen, L, MacKenzie, A.K, Pope, P.B, Chylenski, P, Leiros, I, Williamson, A.K, Christopeit, T, Ostby, H, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome.
PLoS ONE, 13, 2018
|
|
