4G4M
 
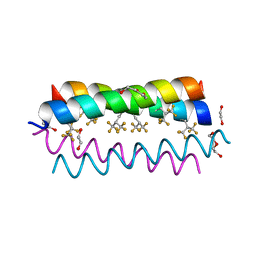 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3(6-13) | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, alpha4F3(6-13) | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4G3B
 
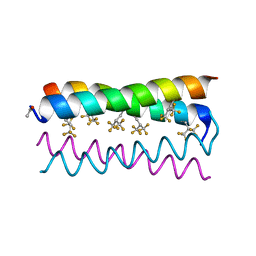 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3d | | Descriptor: | ACETYL GROUP, alpha4F3d | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4G4L
 
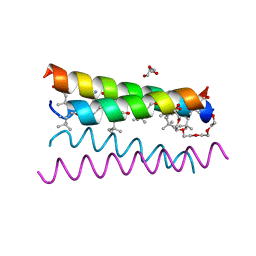 | | Crystal structure of the de novo designed peptide alpha4tbA6 | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
6U91
 
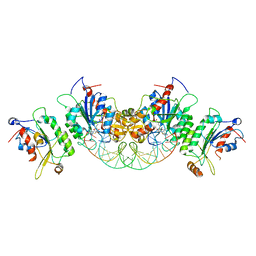 | | Crystal structure of DNMT3B(Q772R)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99998879 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms
To Be Published
|
|
6U90
 
 | | Crystal structure of DNMT3B(N779A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.00081754 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms
To Be Published
|
|
5LKA
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
8CTO
 
 | |
5LY2
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Macrocyclic PEPTIDE Inhibitor CP2_R6Kme3 (13-mer) | | Descriptor: | CHLORIDE ION, CP2_R6Kme3, GLYCEROL, ... | | Authors: | Chowdhury, R, Madden, S.K, Hopkinson, R, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
7UCP
 
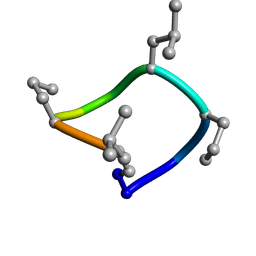 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
8E1E
 
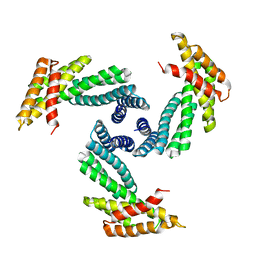 | |
7T2F
 
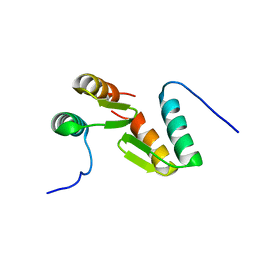 | | Solution structure of the model HEEH mini protein homodimer HEEH_TK_rd5_0341 | | Descriptor: | HEEH mini protein HEEH_TK_rd5_0341 | | Authors: | Lemak, A, Houliston, S, Kim, T.-E, Martel, C, Rocklin, G.J, Arrowsmith, C.H. | | Deposit date: | 2021-12-04 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CUN
 
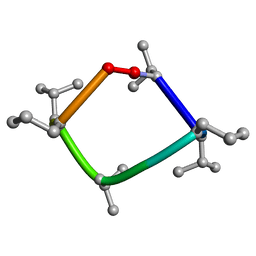 | |
8CWA
 
 | |
8P6I
 
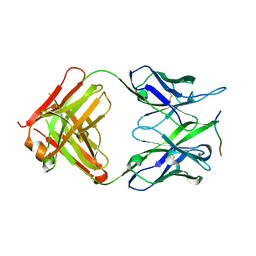 | | Crystal structure of the 139H2 Fab fragment bound to Muc1 peptide epitope | | Descriptor: | 139H2 HC, 139H2 LC, Mucin-1 | | Authors: | Beugelink, J.W, Peng, W, Siborova, M, Pronker, M.F, Snijder, J, Janssen, B.J.C. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing.
Life Sci Alliance, 7, 2024
|
|
5LY1
 
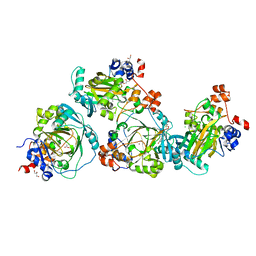 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | Descriptor: | CHLORIDE ION, CP2, GLYCEROL, ... | | Authors: | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
7BAY
 
 | |
3Q7X
 
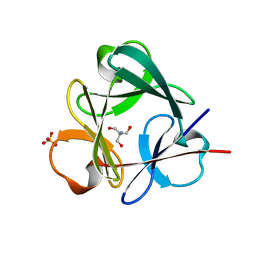 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | Authors: | Blaber, M, Lee, J. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
8DOA
 
 | |
4QCC
 
 | |
7WCQ
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
1OBG
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
