3NFW
 
 | |
2Z1S
 
 | | Beta-glucosidase B from paenibacillus polymyxa complexed with cellotetraose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Isorna, P, Sanz-Aparicio, J. | | Deposit date: | 2007-05-12 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
1F11
 
 | | F124 FAB FRAGMENT FROM A MONOCLONAL ANTI-PRES2 ANTIBODY | | Descriptor: | F124 IMMUNOGLOBULIN (IGG1 HEAVY CHAIN), F124 IMMUNOGLOBULIN (KAPPA LIGHT CHAIN) | | Authors: | Saul, F.A, Vulliez-Le Normand, B, Passafiume, M, Riottot, M.M, Bentley, G.A. | | Deposit date: | 2000-05-18 | | Release date: | 2000-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1SRZ
 
 | | Solution structure of the second complement control protein (CCP) module of the GABA(B)R1a receptor, Pro-119 trans conformer | | Descriptor: | Gamma-aminobutyric acid type B receptor, subunit 1 | | Authors: | Blein, S, Uhrin, D, Smith, B.O, White, J.H, Barlow, P.N. | | Deposit date: | 2004-03-23 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Complement Control Protein (CCP) Modules of GABAB Receptor 1a: ONLY ONE OF THE TWO CCP MODULES IS COMPACTLY FOLDED
J.Biol.Chem., 279, 2004
|
|
3KBD
 
 | | MUTATED NF KAPPA-B SITE, BI MODEL | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*GP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*CP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NF-kappa B binding mechanism: a nuclear magnetic resonance and modeling study of a GGG --> CTC mutation.
Biochemistry, 38, 1999
|
|
2KBD
 
 | | 5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3' | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hantz, E, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-12-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a non-palindromic 16 base-pair DNA related to the HIV-1 kappa B site: evidence for BI-BII equilibrium inducing a global dynamic curvature of the duplex.
J.Mol.Biol., 279, 1998
|
|
7BIZ
 
 | |
4HYP
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | DNA gyrase subunit B, MAGNESIUM ION, N-[7-(1H-imidazol-1-yl)-2-(pyridin-3-yl)[1,3]thiazolo[5,4-d]pyrimidin-5-yl]cyclopropanecarboxamide | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IHO
 
 | | Crystal structure of H-2Db Y159F in complex with chimeric gp100 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Uchtenhagen, H, Stahl, E, Achour, A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Proline substitution independently enhances H-2D(b) complex stabilization and TCR recognition of melanoma-associated peptides
Eur.J.Immunol., 43, 2013
|
|
4J6J
 
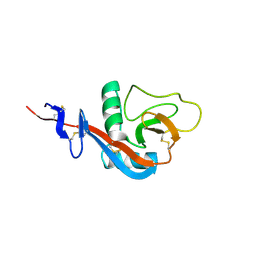 | |
1E15
 
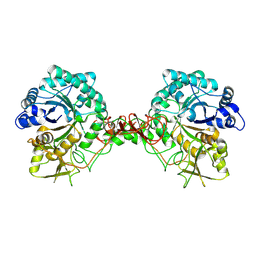 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3ZCV
 
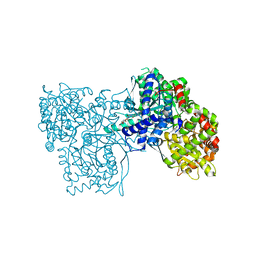 | | Rabbit muscle glycogen phosphorylase b in complex with N-(indol-2- carbonyl)-N-beta-D-glucopyranosyl urea determined at 1.8 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(1H-indol-2-ylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
4J6N
 
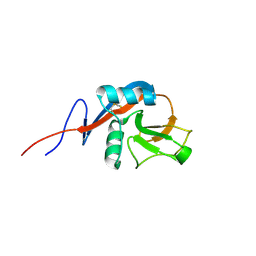 | |
1GPN
 
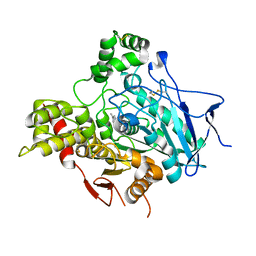 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH HUPERZINE B AT 2.35A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, HUPERZINE B | | Authors: | Dvir, H, Harel, M, Jiang, H.L, Silman, I, Sussman, J.L. | | Deposit date: | 2001-11-07 | | Release date: | 2002-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-Ray Structures of Torpedo Californica Acetylcholinesterase Complexed with (+)-Huperzine a and (-)-Huperzine B: Structural Evidence for an Active Site Rearrangement
Biochemistry, 41, 2002
|
|
1JGE
 
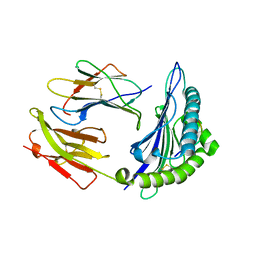 | | HLA-B*2705 bound to nona-peptide m9 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-27 B*2705 ALPHA CHAIN, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-06-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HLA-B27 subtypes differentially associated with disease exhibit subtle structural
alterations
J.Biol.Chem., 277, 2002
|
|
1W1P
 
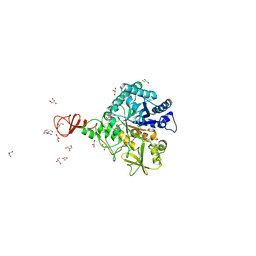 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(Gly-L-Pro) at 2.1 A resolution | | Descriptor: | CHITINASE B, CYCLO-(GLYCINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1W1Y
 
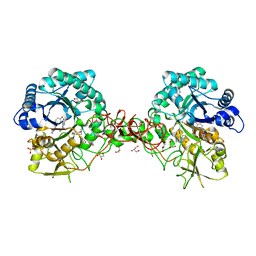 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Tyr-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-TYROSINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
1W1V
 
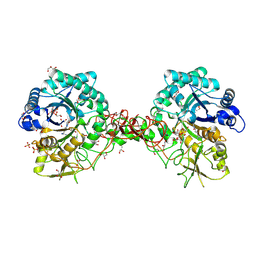 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Arg-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-ARGININE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
3O84
 
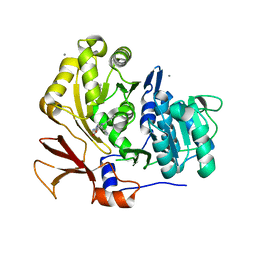 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3BHU
 
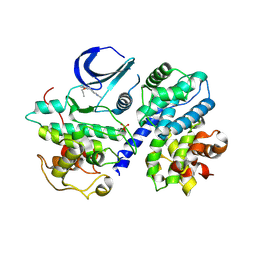 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 5 | | Descriptor: | 4-(4-propoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
2F9B
 
 | | Discovery of Novel Heterocyclic Factor VIIa Inhibitors | | Descriptor: | Coagulation factor VII, Tissue factor, {5-(5-AMINO-1H-PYRROLO[3,2-B]PYRIDIN-2-YL)-6-HYDROXY-3'-NITRO-BIPHENYL-3-YL]-ACETIC ACID | | Authors: | Rai, R, Kolesnikov, A, Sprengeler, P.A, Torkelson, S, Ton, T, Katz, B.A, Yu, C, Hendrix, J, Shrader, W.D, Stephens, R, Cabuslay, R, Sanford, E, Young, W.B. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of novel heterocyclic factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1O4Z
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF BETA-AGARASE B FROM ZOBELLIA GALACTANIVORANS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Allouch, J, Jam, M, Helbert, W, Barbeyron, T, Kloareg, B, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Two {beta}-Agarases.
J.Biol.Chem., 278, 2003
|
|
3ZCQ
 
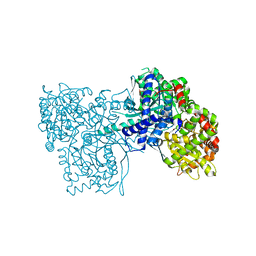 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4- trifluoromethyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2. 15 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-{[4-(trifluoromethyl)benzoyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCR
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4-tert- butyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCT
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(2-naphthoyl) -N-beta-D-glucopyranosyl urea determined at 2.0 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
