5B0Y
 
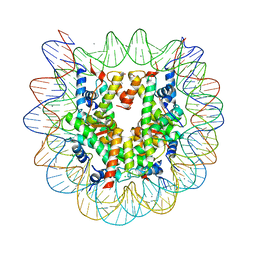 | | Crystal structure of the nucleosome containing histone H3 with the crotonylated lysine 122 | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Suzuki, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5FB0
 
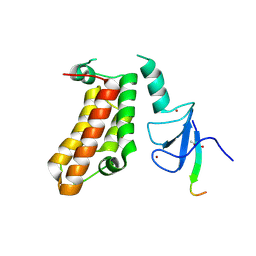 | | Crystal Structure of a PHD finger bound to histone H3 T3ph peptide | | Descriptor: | Nuclear autoantigen Sp-100, Peptide from Histone H3, ZINC ION | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5BNX
 
 | |
5BS7
 
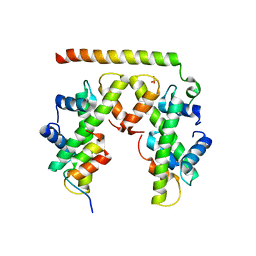 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
2H6N
 
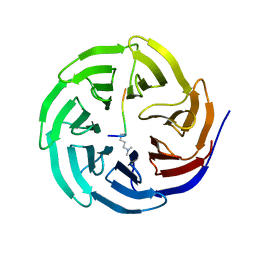 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me2 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5BNV
 
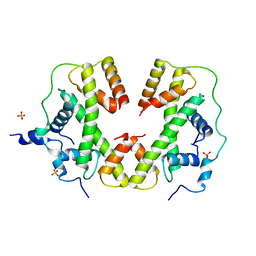 | |
2PVC
 
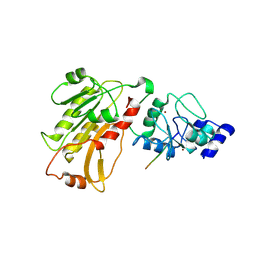 | | DNMT3L recognizes unmethylated histone H3 lysine 4 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, Histone H3 peptide, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
2G46
 
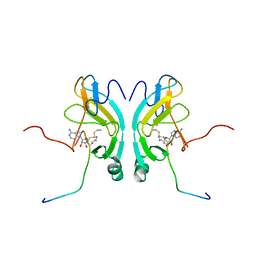 | |
2H6K
 
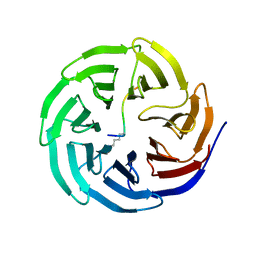 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H13
 
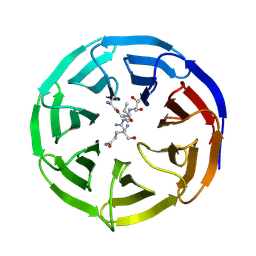 | |
4GNF
 
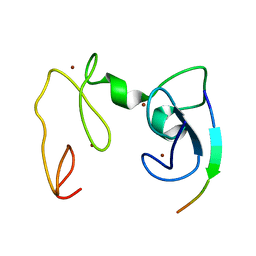 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNE
 
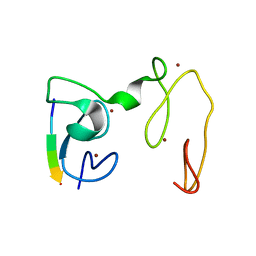 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
5HH7
 
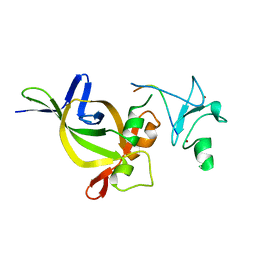 | |
3T6R
 
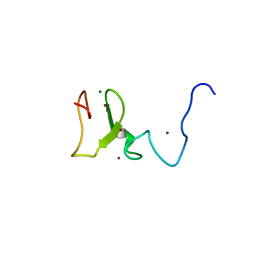 | | Structure of UHRF1 in complex with unmodified H3 N-terminal tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.1t N-terminal peptide, MAGNESIUM ION, ... | | Authors: | Xie, S, Jakoncic, J, Qian, C.M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
J.Mol.Biol., 415, 2012
|
|
3UEE
 
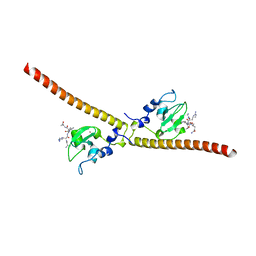 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UED
 
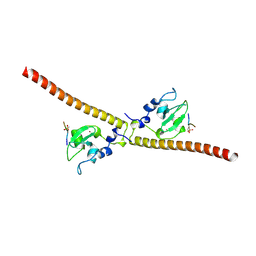 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
4MZH
 
 | |
4MZF
 
 | | Crystal structure of human Spindlin1 bound to histone H3(K4me3-R8me2a) peptide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Peptide from Histone H3.2, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
3UEC
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3PSL
 
 | | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3 based peptide mimetic | | Descriptor: | N-alpha acetylated form of histone H3, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Voronova, A, Skerjanc, I, Couture, J.-F. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic.
Faseb J., 25, 2011
|
|
6DS6
 
 | |
4BD3
 
 | |
3UEF
 
 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
4A0J
 
 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0N
 
 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
