9EMA
 
 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
4JHP
 
 | | The crystal structure of the RPGR RCC1-like domain in complex with PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
7LQ4
 
 | | Rr (RsiG)2-(c-di-GMP)2-WhiG complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), RsiG, WhiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LQ2
 
 | | Apo Rr RsiG- crystal form 1 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, RR RsiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LQ3
 
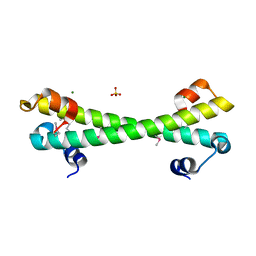 | |
2Q5E
 
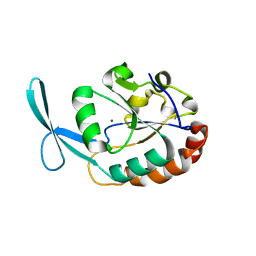 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
7EA8
 
 | |
4JHN
 
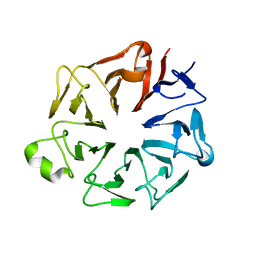 | | The crystal structure of the RPGR RCC1-like domain | | Descriptor: | X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
4OJK
 
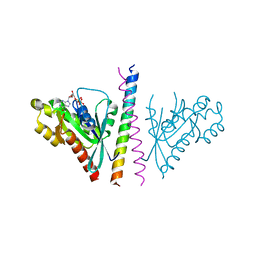 | | Structure of the cGMP Dependent Protein Kinase II and Rab11b Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11B, cGMP-dependent protein kinase 2 | | Authors: | Reger, A.S, Yang, M.P, Guo, E, Kim, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Crystal Structure of the cGMP-dependent Protein Kinase II Leucine Zipper and Rab11b Protein Complex Reveals Molecular Details of G-kinase-specific Interactions.
J.Biol.Chem., 289, 2014
|
|
2JWZ
 
 | | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins | | Descriptor: | Ubiquitin | | Authors: | Haririnia, A, Verma, R, Purohit, N, Twarog, M, Deshaies, R, Bolon, D, Fushman, D. | | Deposit date: | 2007-10-31 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins.
J.Mol.Biol., 375, 2008
|
|
2K2D
 
 | | Solution NMR structure of C-terminal domain of human pirh2. Northeast Structural Genomics Consortium (NESG) target HT2C | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, ZINC ION | | Authors: | Lemak, A, Sheng, Y, Karra, M, Srisailam, S, Laister, R.C, Duan, S, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of Pirh2-mediated p53 ubiquitylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4QHT
 
 | | Crystal structure of AAA+/ sigma 54 activator domain of the flagellar regulatory protein FlrC from Vibrio cholerae in ATP analog bound state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C, MAGNESIUM ION, ... | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
2K2C
 
 | | Solution NMR structure of N-terminal domain of human pirh2. Northeast Structural Genomics Consortium (NESG) target HT2A | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, ZINC ION | | Authors: | Wu, B, Lemak, A, Sheng, Y, Karra, M, Srisailam, S, Sunnerhagen, M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of Pirh2-mediated p53 ubiquitylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6ZQC
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Pre-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQA
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4PR6
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
6ZQB
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
2KOJ
 
 | |
4QHS
 
 | | Crystal structure of AAA+sigma 54 activator domain of the flagellar regulatory protein FlrC of Vibrio cholerae in nucleotide free state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
2K6Q
 
 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
2GHQ
 
 | |
4PRF
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
