5NEG
 
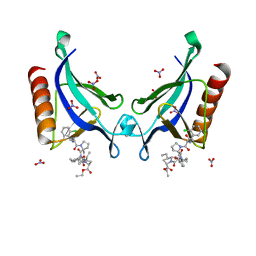 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-13]-OEt | | Descriptor: | NITRATE ION, Protein enabled homolog, SULFATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5H2N
 
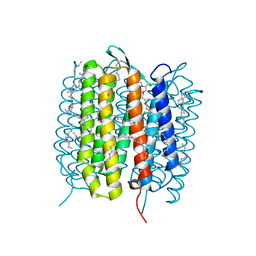 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5UER
 
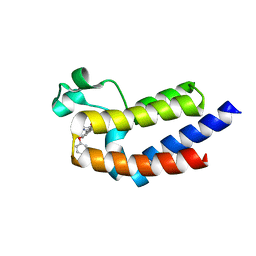 | | BRD4_BD2_A-1359930 | | Descriptor: | Bromodomain-containing protein 4, N-[3-(3-methyl-4-oxo-4,5,6,7-tetrahydro-2H-isoindol-1-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Complex structure of BRD4_BD2_A-1359930
To Be Published
|
|
5H2O
 
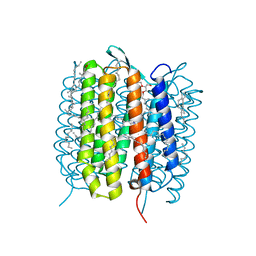 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
4C2Y
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
5TIX
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/R-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2R)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5Q0Q
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl 4-({2-phenyl-5-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine-3-carbonyl}amino)benzoate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
3AOD
 
 | | Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, RIFAMPICIN | | Authors: | Nakashima, R, Sakurai, K, Yamaguchi, A. | | Deposit date: | 2010-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket
Nature, 480, 2011
|
|
3OH3
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE -Arabinose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2S,3R,4S,5S)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OFS
 
 | | Dynamic conformations of the CD38-mediated NAD cyclization captured using multi-copy crystallography | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Conformations of the CD38-Mediated NAD Cyclization Captured in a Single Crystal
J.Mol.Biol., 405, 2011
|
|
4RKU
 
 | | Crystal structure of plant Photosystem I at 3 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Mazor, Y, Borovikova, A, Greenberg, I, Nelson, N. | | Deposit date: | 2014-10-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant Photosystem I at 3.1 Angstrom resolution
To be Published
|
|
4GS6
 
 | | Irreversible Inhibition of TAK1 Kinase by 5Z-7-Oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, 1,2-ETHANEDIOL, Tak1-Tab1 fusion protein | | Authors: | Larsen, N.A, Ferguson, A.D, Wu, J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism and In Vitro Pharmacology of TAK1 Inhibition by (5Z)-7-Oxozeaenol.
Acs Chem.Biol., 8, 2013
|
|
5POX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11075a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
5UEP
 
 | | BRD4_BD2_A-581577 | | Descriptor: | Bromodomain-containing protein 4, ethyl 3-methyl-4-oxo-4,5,6,7-tetrahydro-2H-isoindole-1-carboxylate | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex structure of BRD4_BD2_A-581577
To Be Published
|
|
3JX4
 
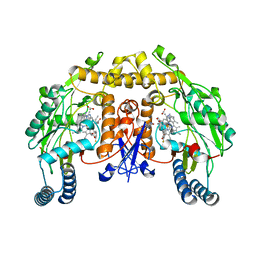 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3RIF
 
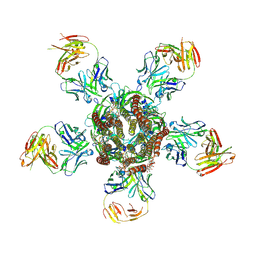 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate. | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
5U6B
 
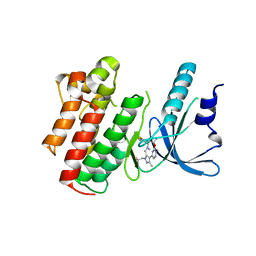 | | Structure of the Axl kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase receptor UFO | | Authors: | Gajiwala, K.S, Grodsky, N. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
5CEL
 
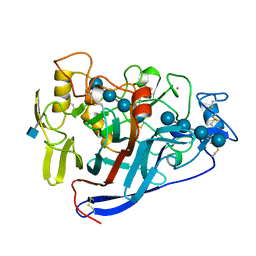 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
5TB6
 
 | | Structure of bromodomain of CREBBP with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
4OYT
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with D-serine and folinic acid | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-02-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5PNZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10162a | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-methoxyphenyl)methyl]-1H-tetrazole, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5T8H
 
 | |
3WGV
 
 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state with oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
4KWP
 
 | | Crystal Structure of Human CK2-alpha in complex with a benzimidazole inhibitor (K164) at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-tetrabromo-1-(2-deoxy-beta-D-erythro-pentofuranosyl)-1H-benzimidazole, Casein kinase II subunit alpha, ... | | Authors: | Ranchio, A, Lolli, G, Battistutta, R. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: structural features and pharmacological potential.
Cell.Mol.Life Sci., 71, 2014
|
|
