6M4H
 
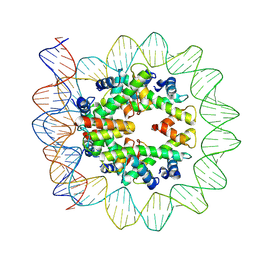 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (103-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6M44
 
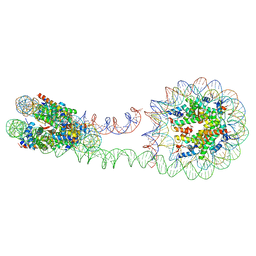 | | 355 bp di-nucleosome harboring cohesive DNA termini (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
4JPG
 
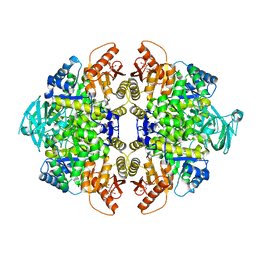 | | 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as Novel PKM2 Activators | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-(1H-benzimidazol-1-ylmethyl)-4H-pyrido[1,2-a]pyrimidin-4-one, Pyruvate kinase isozymes M1/M2 | | Authors: | Greasley, S.E, Hickey, M, Phonephaly, H, Cronin, C. | | Deposit date: | 2013-03-19 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as novel PKM2 activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4QG9
 
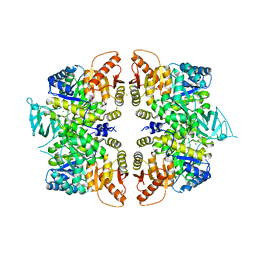 | | crystal structure of PKM2-R399E mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4H9N
 
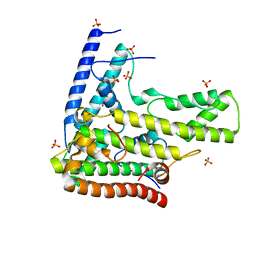 | | Complex structure 1 of DAXX/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4H9P
 
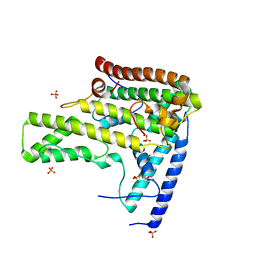 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4F6N
 
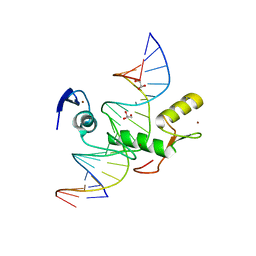 | | Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*GP*AP*(5CM)P*GP*(5CM)P*GP*GP*TP*GP*AP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*TP*CP*AP*CP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*AP*TP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F6M
 
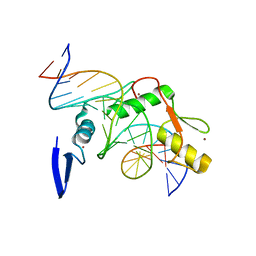 | | Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OQA
 
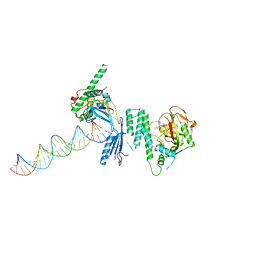 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4F3L
 
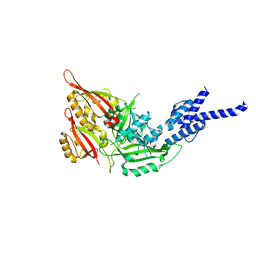 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
2P1B
 
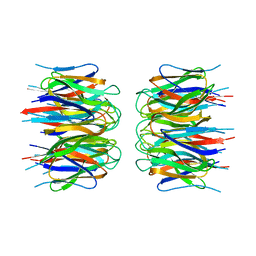 | | Crystal structure of human nucleophosmin-core | | Descriptor: | Nucleophosmin | | Authors: | Lee, H.H, Kim, H.S, Kang, J.Y, Lee, B.I, Ha, J.Y, Yoon, H.J, Lim, S.O, Jung, G, Suh, S.W. | | Deposit date: | 2007-03-03 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface
Proteins, 69, 2007
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6WP4
 
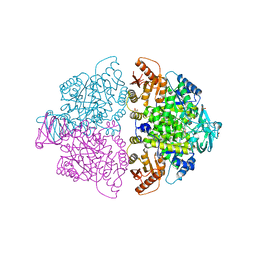 | | Pyruvate Kinase M2 mutant-S37E | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nandi, S, Razzaghi, M, Srivastava, D, Dey, M. | | Deposit date: | 2020-04-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation.
J.Biol.Chem., 295, 2020
|
|
6R0C
 
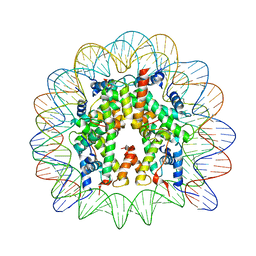 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | Descriptor: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
6R94
 
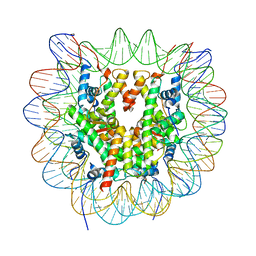 | | Cryo-EM structure of NCP_THF2(-3) | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
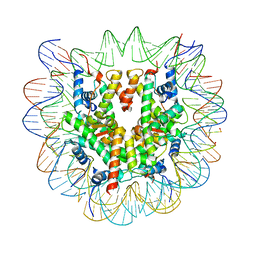 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6XTY
 
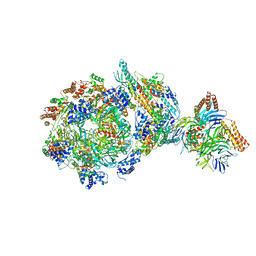 | | CryoEM structure of human CMG bound to AND-1 (CMGA) | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Rzechorzek, N.J, Pellegrini, L, Chirgadze, D.Y, Hardwick, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
6RO1
 
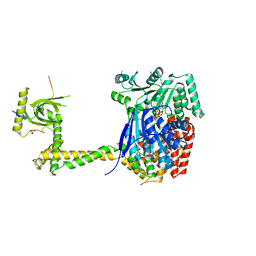 | | X-ray crystal structure of the MTR4 NVL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Lingaraju, M, Langer, L.M, Basquin, J, Falk, S, Conti, E. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Nat Commun, 10, 2019
|
|
6RO4
 
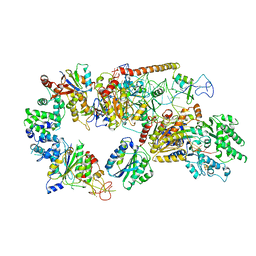 | | Structure of the core TFIIH-XPA-DNA complex | | Descriptor: | DNA repair protein complementing XP-A cells, DNA1, DNA2, ... | | Authors: | Kokic, G, Chernev, A, Tegunov, D, Dienemann, C, Urlaub, H, Cramer, P. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of TFIIH activation for nucleotide excision repair.
Nat Commun, 10, 2019
|
|
6RXS
 
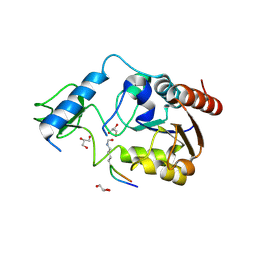 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YOV
 
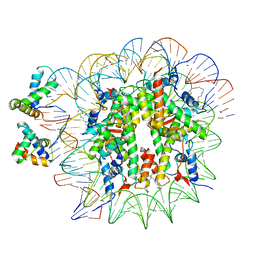 | | OCT4-SOX2-bound nucleosome - SHL+6 | | Descriptor: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6SEG
 
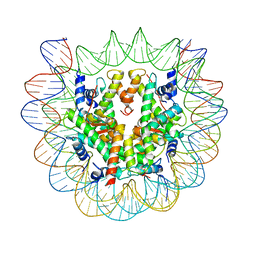 | | Class1: CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
6SE0
 
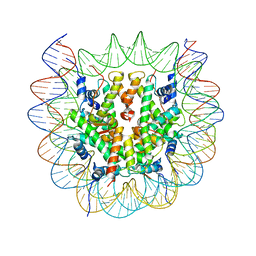 | | Class 1 : CENP-A nucleosome | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
6YAN
 
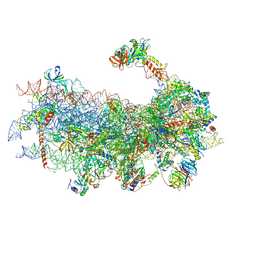 | | Mammalian 48S late-stage translation initiation complex with histone 4 mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
