5J8U
 
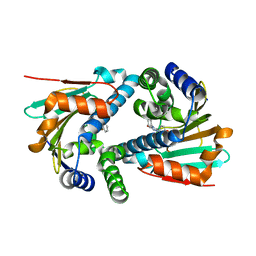 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5FDO
 
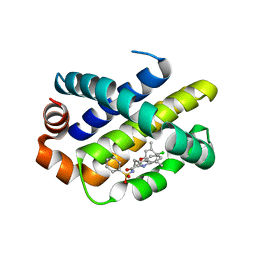 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-~{N}-(phenylsulfonyl)-1~{H}-indole-2-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
3DFN
 
 | |
5FE0
 
 | | Crystal structure of human PCAF bromodomain in complex with acetyllysine | | Descriptor: | Histone acetyltransferase KAT2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5JMB
 
 | |
8HLO
 
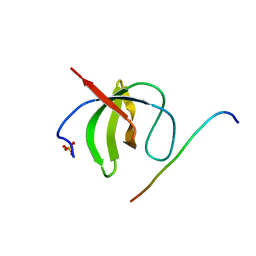 | | Crystal structure of ASAP1-SH3 and MICAL1-PRM complex | | Descriptor: | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, Proline rich motif from MICAL1, ... | | Authors: | Niu, F, Wei, Z, Yu, C. | | Deposit date: | 2022-11-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Int J Mol Sci, 24, 2023
|
|
8HTW
 
 | |
3D8K
 
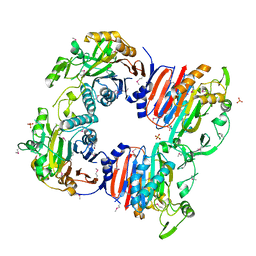 | |
5FST
 
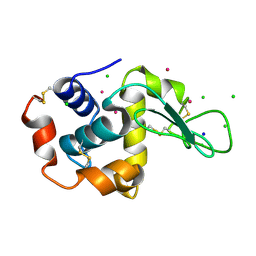 | | Structure of lysozyme prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME C, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, P, Carpentier, P. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
5FTS
 
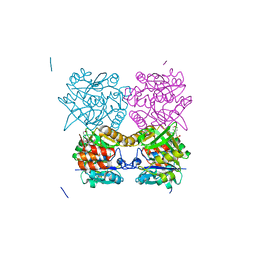 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
5FPN
 
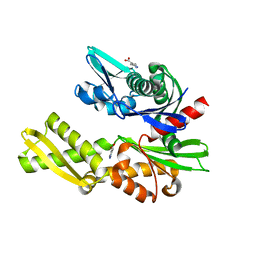 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 3,5-dimethyl-1H-pyrazole-4-carboxylic acid (AT9084) in an alternate binding site. | | Descriptor: | 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, HEAT SHOCK-RELATED 70 KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3DA2
 
 | | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 13, N-(4-chlorobenzyl)-N-methylbenzene-1,4-disulfonamide, ... | | Authors: | Pilka, E.S, Picaud, S.S, Yue, W.W, King, O.N.F, Bray, J.E, Filippakopoulos, P, Roos, A.K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor.
To be Published
|
|
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8SM1
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Immunity, 58, 2025
|
|
5JOG
 
 | | CRYSTAL STRUCTURE OF CSN5(2-257) IN COMPLEX WITH CNS5i-3 | | Descriptor: | 3-(difluoromethyl)-N-{6-[(5S,6S)-6-hydroxy-6,7,8,9-tetrahydro-5H-imidazo[1,5-a]azepin-5-yl][1,1'-biphenyl]-3-yl}-1-(propan-2-yl)-1H-pyrazole-5-carboxamide, COP9 signalosome complex subunit 5, ZINC ION | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Targeted inhibition of the COP9 signalosome for treatment of cancer.
Nat Commun, 7, 2016
|
|
5F8V
 
 | |
5F9L
 
 | |
8HE0
 
 | |
3DB8
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 041 | | Descriptor: | 3-[3-chloro-5-(5-{[(1S)-1-phenylethyl]amino}isoxazolo[5,4-c]pyridin-3-yl)phenyl]propan-1-ol, Polo-like kinase 1 | | Authors: | Elling, R.A, Hanan, E.J, Lew, W, Romanowski, M.J. | | Deposit date: | 2008-05-30 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design and synthesis of 2-amino-isoxazolopyridines as Polo-like kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBL
 
 | |
5IUM
 
 | | Crystal structure of phosphorylated DesKC | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
8HFB
 
 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Eom, H, Song, W.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
3D8A
 
 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
5FYZ
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 2-(2-oxo-2,3-dihydro-1H-indol-3-yl)acetonitrile (N10063a) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N10063A
To be Published
|
|
5FA7
 
 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
