6Y53
 
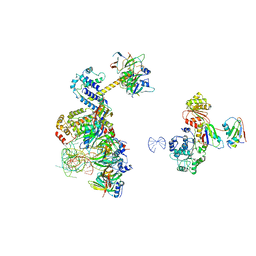 | | human 17S U2 snRNP low resolution part | | Descriptor: | HIV Tat-specific factor 1, Probable ATP-dependent RNA helicase DDX46, Small nuclear ribonucleoprotein E, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6PDT
 
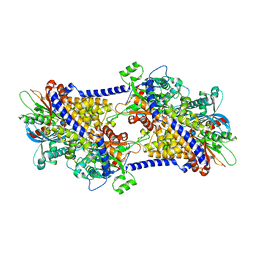 | | cryoEM structure of yeast glucokinase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucokinase-1, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dosey, A.M, Farrell, D.P, Stoddard, P.R, Kollman, J.M. | | Deposit date: | 2019-06-19 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Polymerization in the actin ATPase clan regulates hexokinase activity in yeast.
Science, 367, 2020
|
|
6S7M
 
 | |
6P9I
 
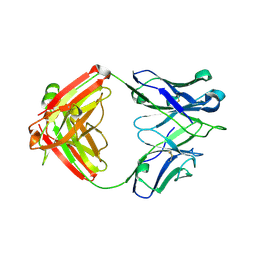 | | crystal structure of human anti staphylococcus aureus antibody STAU-399 Fab | | Descriptor: | human anti staphylococcus aureus antibody STAU-399 Fab heavy chain, human anti staphylococcus aureus antibody STAU-399 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6PJE
 
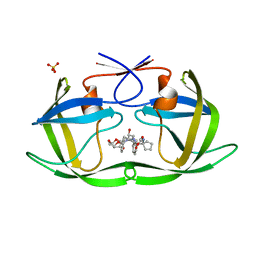 | | HIV-1 Protease NL4-3 WT in Complex with LR2-43 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJO
 
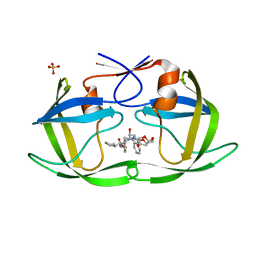 | | HIV-1 Protease NL4-3 WT in Complex with LR2-42 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopentyl-2-({(2S,4S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-4-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6S4I
 
 | |
6P3D
 
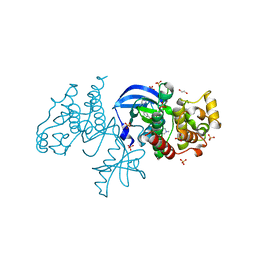 | | The co-crystal structure of BRAF(V600E) with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, AMMONIUM ION, ... | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-05-23 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
8BZC
 
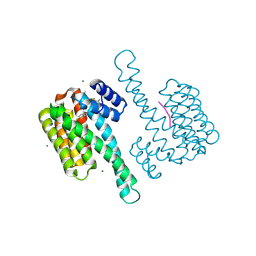 | |
6TXE
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dTMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
8C41
 
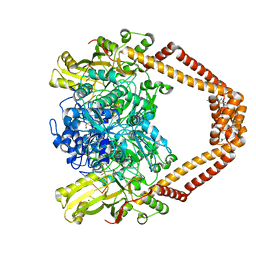 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*GP*AP*AP*T)-3'), ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8FDP
 
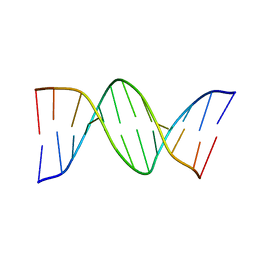 | |
7ZRC
 
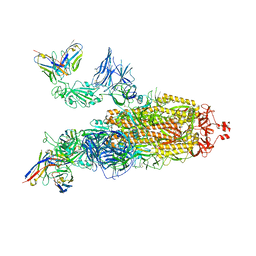 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab Heavy Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-04 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
6YDX
 
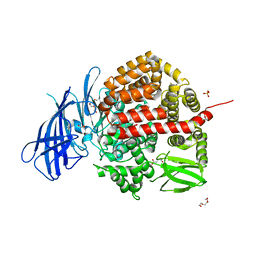 | | Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[[(4~{R},8~{S},11~{S})-11-azanyl-8-[(4-hydroxyphenyl)methyl]-6,10-bis(oxidanylidene)-1,2-dithia-5,9-diazacyclotridec-4-yl]carbonylamino]methyl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Saridakis, E, Giastas, P, Stratikos, E. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6YHN
 
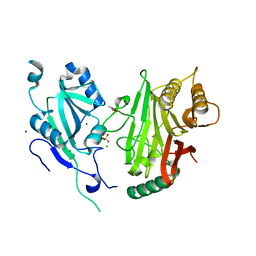 | | Crystal structure of domains 4-5 of CNFy from Yersinia pseudotuberculosis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Cytotoxic necrotizing factor, ... | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
7SZO
 
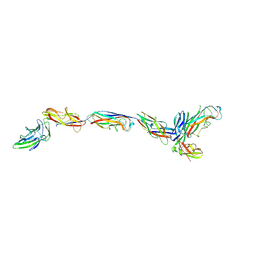 | | Structure of a bacterial fimbrial tip containing FocH | | Descriptor: | Chaperone protein FimC, FimF protein, FimG, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Aprikian, P, Sokurenko, E.V. | | Deposit date: | 2021-11-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant FimH Adhesin Demonstrates How the Allosteric Catch Bond Mechanism Can Support Fast and Strong Bacterial Attachment in the Absence of Shear.
J.Mol.Biol., 434, 2022
|
|
6F3L
 
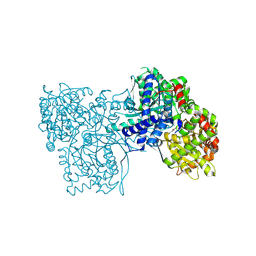 | | The crystal structure of Glycogen Phosphorylase in complex with 10b | | Descriptor: | 6-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-1~{H}-1,2,4-triazol-3-yl]naphthalene-2-carboxylic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6PJH
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-28 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PM9
 
 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
6PGL
 
 | | Structure of Kluyveromyces marxianus Usb1 with uridine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, GLYCEROL, ... | | Authors: | Nomura, Y, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis for the evolution of cyclic phosphodiesterase activity in the U6 snRNA exoribonuclease Usb1.
Nucleic Acids Res., 48, 2020
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
6PPQ
 
 | |
6PJF
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-44 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopentyl-2-({(2S,3S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
4Y8H
 
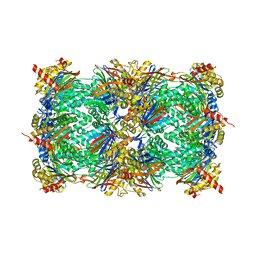 | | Yeast 20S proteasome in complex with N3-APAL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
