7TLC
 
 | |
7TP7
 
 | |
7TL9
 
 | |
7TL1
 
 | |
7TGE
 
 | |
7RZU
 
 | |
7RZS
 
 | |
7RZR
 
 | |
7RZT
 
 | |
7RZV
 
 | |
7RZQ
 
 | |
7TUZ
 
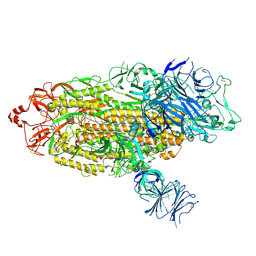
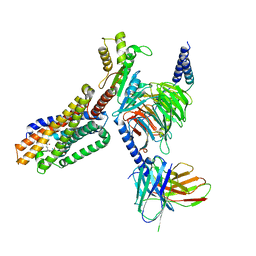 | | Cryo-EM structure of 7alpha,25-dihydroxycholesterol-bound EBI2/GPR183 in complex with Gi protein | | Descriptor: | (2S,4aS,4bS,7R,8S,8aS,9R,10aR)-7-[(2R,3R)-7-hydroxy-3,7-dimethyloctan-2-yl]-4a,7,8-trimethyltetradecahydrophenanthrene-2,9-diol, G-protein coupled receptor 183, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, H, Hung, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7TUY
 
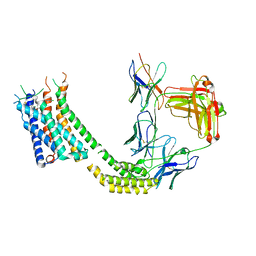 | | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | | Descriptor: | 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, G-protein coupled receptor 183,Soluble cytochrome b562 fusion, anti-BRIL Fab Heavy chain, ... | | Authors: | Chen, H, Huang, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7VFL
 
 | | Crystal structure of SdgB (UDP, NAG, and O-glycosylated SD peptide-binding form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFO
 
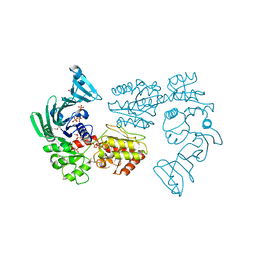 | | Crystal structure of SdgB (Phosphate-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFM
 
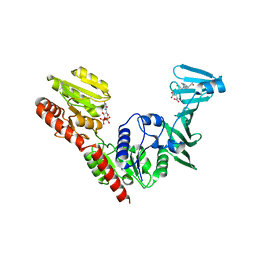 | | Crystal structure of SdgB (UDP and SD peptide-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, SER-ASP-SER-ASP, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFN
 
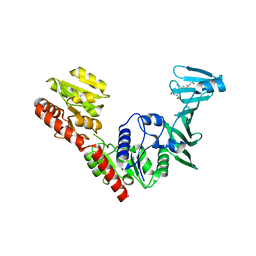 | | Crystal structure of SdgB (SD peptide-binding form) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFK
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VEN
 
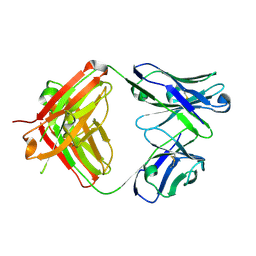 | | Structure of burosumab Fab | | Descriptor: | Burosumab Fab HC, Burosumab Fab LC | | Authors: | Heo, Y.S. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Fab Fragment of Burosumab
To Be Published
|
|
7UL3
 
 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL5
 
 | |
7UL2
 
 | | CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Nanobody 6, Neurotensin receptor 1, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UYI
 
 | |
7UMN
 
 | |
