6G4Z
 
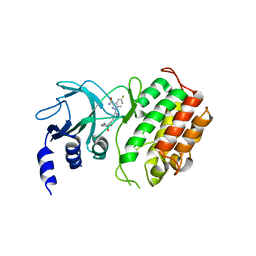 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | | Descriptor: | 5-fluoranyl-1-[4-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrol-3-yl]ethynyl]pyridin-2-yl]indazole-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Leonardo-Silvestre, H, McEwan, P.A, Hymowitz, S.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
7SCO
 
 | | Structure of H1 influenza hemagglutinin bound to Fab 310-39G10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 310-39G10 Fab, ... | | Authors: | Torrents de la Pena, A, Ward, A.B. | | Deposit date: | 2021-09-28 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Allelic polymorphism controls autoreactivity and vaccine elicitation of human broadly neutralizing antibodies against influenza virus.
Immunity, 55, 2022
|
|
7SCN
 
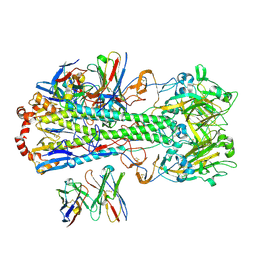 | |
6G4Y
 
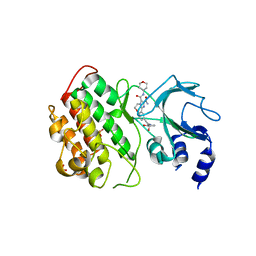 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 1a | | Descriptor: | 10-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrolidin-3-yl]ethynyl]-~{N}3-(oxan-4-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Hole, A.J, Hymowitz, S.G, McEwan, P.A. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
7SZR
 
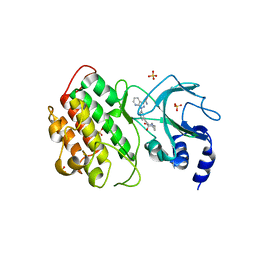 | | NIK bound to inhibitor G02792917 | | Descriptor: | 1-(3-{[(1R,4R,5S)-4-hydroxy-2-methyl-3-oxo-2-azabicyclo[3.1.0]hexan-4-yl]ethynyl}phenyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Liau, N.P.D, Hymowitz, S.G. | | Deposit date: | 2021-11-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a nick in NIK: Extending the half-life of a NIK inhibitor through structure-based drug design.
Bioorg.Med.Chem.Lett., 89, 2023
|
|
7M61
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 1 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M64
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M62
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M65
 
 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils seeded by patient extracted fibrils, polymorph 4 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores
Nat.Struct.Mol.Biol., 28, 2021
|
|
8QPT
 
 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
2Y3J
 
 | | Structure of segment AIIGLM from the amyloid-beta peptide (Ab, residues 30-35) | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7ZHL
 
 | | Salmonella enterica Rhs1 C-terminal toxin TreTu | | Descriptor: | RHS repeat protein, ZINC ION | | Authors: | Jurenas, D, Rey, M, Chamot-Rooke, J, Terradot, L, Cascales, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Salmonella antibacterial Rhs polymorphic toxin inhibits translation through ADP-ribosylation of EF-Tu P-loop.
Nucleic Acids Res., 50, 2022
|
|
7ZHM
 
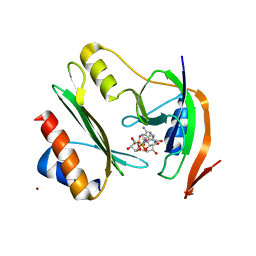 | | Salmonella enterica Rhs1 C-terminal toxin TreTu complex with TriTu immunity protein | | Descriptor: | Immunity protein TriTu, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rhs1 protein, ... | | Authors: | Jurenas, D, Rey, M, Chamot-Rooke, J, Terradot, L, Cascales, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Salmonella antibacterial Rhs polymorphic toxin inhibits translation through ADP-ribosylation of EF-Tu P-loop.
Nucleic Acids Res., 50, 2022
|
|
6EJJ
 
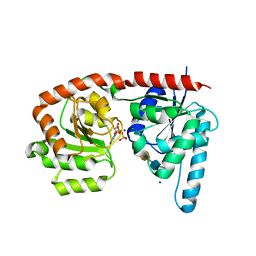 | | Structure of a glycosyltransferase / state 2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, NerylNeryl pyrophosphate, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
8UGC
 
 | |
5ZCT
 
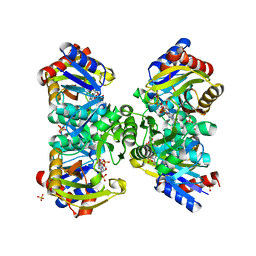 | | The crystal structure of the poly-alpha-L-glutamate peptides synthetase RimK at 2.05 angstrom resolution. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6--L-glutamate ligase, ... | | Authors: | Arimura, Y, Kono, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2018-02-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural polymorphism of the Escherichia coli poly-alpha-L-glutamate synthetase RimK
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6EJI
 
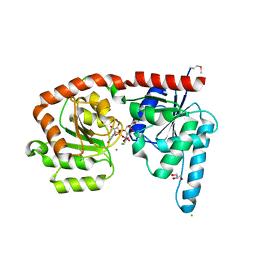 | | Structure of a glycosyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
4WQP
 
 | |
5K6P
 
 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
5KN7
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LQ6
 
 | | Salmonella effector SpvD - R161 variant | | Descriptor: | SODIUM ION, Virulence protein vsdE | | Authors: | Zhang, Y, Grabe, G.J, Rolhion, N, Yang, Y, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
4WPF
 
 | | Crystal structure of RORc in complex with a phenyl sulfonamide agonist | | Descriptor: | N-[4-(4-acetylpiperazin-1-yl)-2-fluorobenzyl]-N-cyclobutylbenzenesulfonamide, Nuclear receptor ROR-gamma, RHKILHRLLQEGSPS | | Authors: | Kiefer, J.R, Wallweber, H.A, de Leon Boenig, G, Hymowitz, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Minor Structural Change to Tertiary Sulfonamide RORc Ligands Led to Opposite Mechanisms of Action.
Acs Med.Chem.Lett., 6, 2015
|
|
6U2H
 
 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1NU7
 
 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
