4P0F
 
 | | Cleaved Serpin 42Da (C 2 2 21) | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
7O1D
 
 | |
7O2L
 
 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
1IR7
 
 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
1IR9
 
 | | IM mutant of lysozyme | | Descriptor: | lysozyme | | Authors: | Ohmura, T, Ueda, T, Hashimoto, Y, Imoto, T. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme.
Protein Eng., 14, 2001
|
|
1PIS
 
 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
3D0M
 
 | | X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3'), SULFATE ION | | Authors: | Rypniewski, W, Adamiak, D.A, Milecki, J, Adamiak, R.W. | | Deposit date: | 2008-05-02 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex.
Rna, 14, 2008
|
|
3CR1
 
 | | crystal structure of a minimal, mutant, all-RNA hairpin ribozyme (A38C, A-1OMA) grown from MgCl2 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3'), RNA (5'-R(*UP*CP*GP*UP*GP*GP*UP*CP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3'), ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
5E1S
 
 | | The Crystal structure of INSR Tyrosine Kinase in complex with the Inhibitor BI 885578 | | Descriptor: | (5R)-N-(1-{2-[4-(2-methoxyethyl)piperazin-1-yl]ethyl}-1H-pyrazol-3-yl)-5,8-dimethyl-9-phenyl-6,8-dihydro-5H-pyrazolo[3,4-h]quinazolin-2-amine, Insulin receptor | | Authors: | Kessler, D, Zahn, S, Sanderson, M, Wolkerstorfer, B. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | BI 885578, a Novel IGF1R/INSR Tyrosine Kinase Inhibitor with Pharmacokinetic Properties That Dissociate Antitumor Efficacy and Perturbation of Glucose Homeostasis.
Mol.Cancer Ther., 14, 2015
|
|
3CZW
 
 | | X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex | | Descriptor: | RNA (5'-R(*G*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3'), SULFATE ION | | Authors: | Rypniewski, W, Adamiak, D.A, Milecki, J, Adamiak, R.W. | | Deposit date: | 2008-04-30 | | Release date: | 2008-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex.
Rna, 14, 2008
|
|
8SSE
 
 | | Methionine synthase, C-terminal fragment, Cobalamin and Reactivation Domains from Thermus thermophilus HB8 | | Descriptor: | COBALAMIN, Methionine synthase | | Authors: | Yamada, K, Mendoza, J, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of full-length cobalamin-dependent methionine synthase and cofactor loading captured in crystallo.
Nat Commun, 14, 2023
|
|
6C0Y
 
 | |
8HEY
 
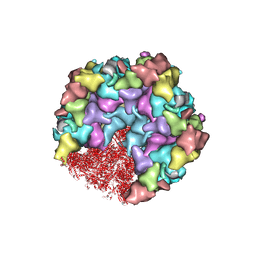 | | One CVSC-binding penton vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEX
 
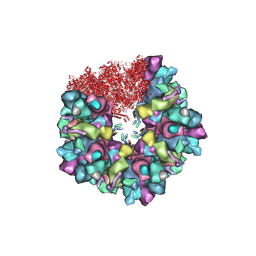 | | C5 portal vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
2BSI
 
 | |
2C0A
 
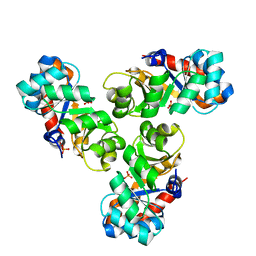 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | KHG/KDPG ALDOLASE, PHOSPHATE ION | | Authors: | Hutchins, M.J, Fullerton, S.W.B, Toone, E.J, Naismith, J.H. | | Deposit date: | 2005-08-29 | | Release date: | 2005-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
2BSH
 
 | |
8HPJ
 
 | |
8I2G
 
 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
2BY4
 
 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the thapsigargin derivative Boc-12ADT. | | Descriptor: | (3S,3AR,4S,6S,6AR,7S,8S,9BS)-6-(ACETYLOXY)-3,3A-DIHYDROXY-3,6,9-TRIMETHYL-8-{[(2Z)-2-METHYLBUT-2-ENOYL]OXY}-7-(OCTANOYLOXY)-2-OXO-2,3,3A,4,5,6,6A,7,8,9B-DECAHYDROAZULENO[4,5-B]FURAN-4-YL 12-[(TERT-BUTOXYCARBONYL)AMINO]DODECANOATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Jensen, A.L, Moller, J.V, Soehoel, H, Denmeade, S.R, Isaacs, J.T, Olsen, C.E, Christensen, S.B, Nissen, P. | | Deposit date: | 2005-07-28 | | Release date: | 2006-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Natural Products as Starting Materials for Development of Second-Generation Serca Inhibitors Targeted Towards Prostate Cancer Cells
Bioorg.Med.Chem., 14, 2006
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
5F74
 
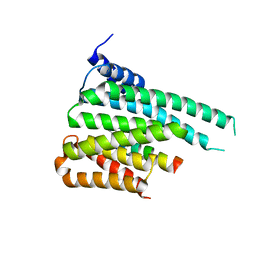 | | Crystal structure of ChREBP:14-3-3 complex bound with AMP | | Descriptor: | 14-3-3 protein beta/alpha, ADENOSINE MONOPHOSPHATE, Carbohydrate-responsive element-binding protein | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR.
J.Biol.Chem., 291, 2016
|
|
4Y5I
 
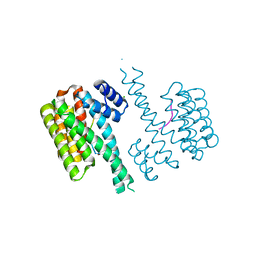 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5HF3
 
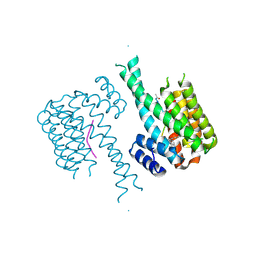 | | Crystal structure of C-terminal modified Tau peptide-hybrid 201D with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, modified Tau peptide | | Authors: | Bartel, M, Milroy, L.G, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
