3PM2
 
 | | Crystal structure of a novel type of odorant binding protein from Anopheles gambiae belonging to the c+ class | | Descriptor: | Odorant binding protein (AGAP007287-PA) | | Authors: | Spinelli, S, Lagarde, A, Qiao, H, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel type of odorant-binding protein from Anopheles gambiae, belonging to the C-plus class.
Biochem.J., 437, 2011
|
|
4TZM
 
 | |
4HEE
 
 | | Crystal structure of PPARgamma in complex with compound 13 | | Descriptor: | 5-benzyl-2-ethyl-3-{(1S)-5-[2-(1H-tetrazol-5-yl)phenyl]-2,3-dihydro-1H-inden-1-yl}-3,5-dihydro-4H-imidazo[4,5-c]pyridin-4-one, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Han, S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and evaluation of imidazo[4,5-c]pyridin-4-one derivatives with dual activity at angiotensin II type 1 receptor and peroxisome proliferator-activated receptor-gamma
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2SRC
 
 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
1TN4
 
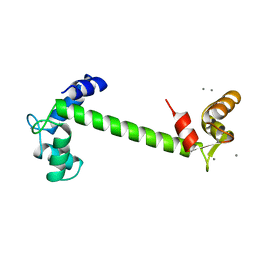 | | FOUR CALCIUM TNC | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | Deposit date: | 1997-09-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
2PX7
 
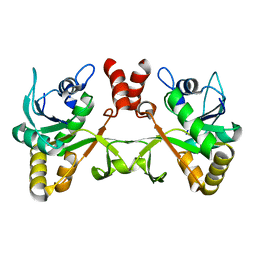 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | | Authors: | Chen, L, Tsukuda, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chen, L.-Q, Liu, Z.-J, Lee, D, Chang, S.-H, Nguyen, D, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8.
To be Published
|
|
2LNM
 
 | |
3QIB
 
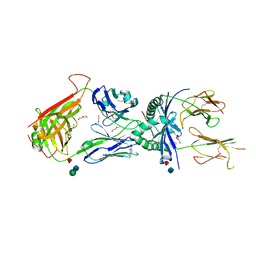 | | Crystal structure of the 2B4 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 beta chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
6BNA
 
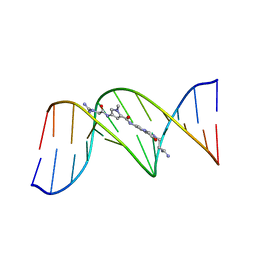 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | Authors: | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | Deposit date: | 1984-08-30 | | Release date: | 1984-10-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
3PTD
 
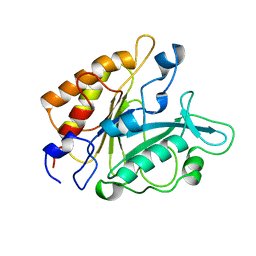 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D274S | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-17 | | Release date: | 1998-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
2XI3
 
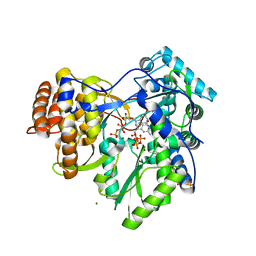 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2LWX
 
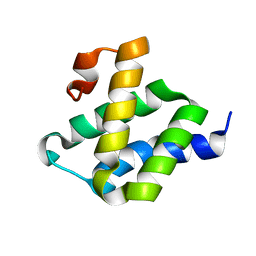 | |
3RC4
 
 | |
3GV4
 
 | | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG | | Descriptor: | CALCIUM ION, Histone deacetylase 6, ZINC ION, ... | | Authors: | Dong, A, Ravichandran, M, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG
To be Published
|
|
5TRR
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
4J1V
 
 | | Functional and structural studies of MOBKL1B, a Salvador/Warts/Hippo tumor suppressor pathway, in HCV replication | | Descriptor: | MOB kinase activator 1A, NS5A domain II peptide, ZINC ION | | Authors: | Chung, H.-Y, Gu, M, Rice, C.M. | | Deposit date: | 2013-02-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Seed Sequence-Matched Controls Reveal Limitations of Small Interfering RNA Knockdown in Functional and Structural Studies of Hepatitis C Virus NS5A-MOBKL1B Interaction.
J.Virol., 88, 2014
|
|
6AO3
 
 | |
5BND
 
 | | Crystal structure of the C-terminal domain of TagH | | Descriptor: | ABC transporter, ATP-binding protein | | Authors: | Chen, S.C, Chen, Y. | | Deposit date: | 2015-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SH3-Like Motif-Containing C-terminal Domain of Staphylococcal Teichoic Acid Transporter Suggests Possible Function.
Proteins, 2016
|
|
1RFA
 
 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | Descriptor: | RAF1 | | Authors: | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | Deposit date: | 1995-04-26 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
1XX0
 
 | | Structure of the C-terminal PH domain of human pleckstrin | | Descriptor: | Pleckstrin | | Authors: | Edlich, C, Stier, G, Simon, B, Sattler, M, Muhle-Goll, C. | | Deposit date: | 2004-11-03 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and phosphatidylinositol-(3,4)-bisphosphate binding of the C-terminal PH domain of human pleckstrin
STRUCTURE, 13, 2005
|
|
2BLM
 
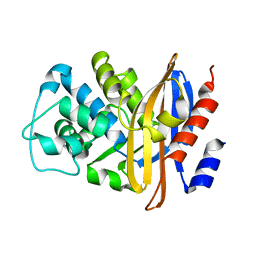 | |
335D
 
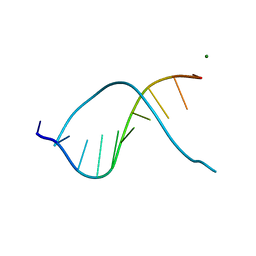 | |
2M45
 
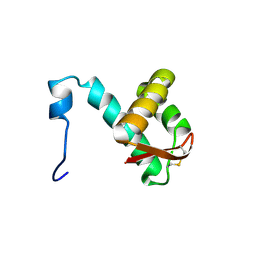 | | NMR solution structure of the C-terminus of the minichromosome maintenance protein MCM from Sulfolobus solfataricus | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Wiedemann, C, Ohlenschlager, O, Medagli, B, Onesti, S, Gorlach, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex
Nucleic Acids Res., 43, 2015
|
|
5TZF
 
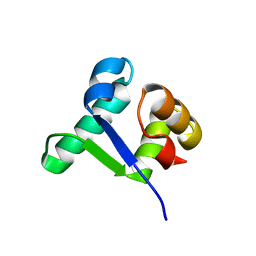 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2 intermediate, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
2EWE
 
 | |
