6GFY
 
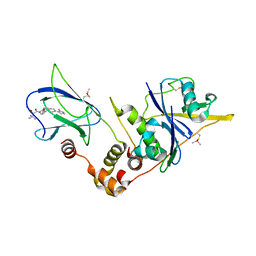 | | pVHL:EloB:EloC in complex with modified VH032 containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 14a) | | Descriptor: | (2~{R},3~{R},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
2YPY
 
 | |
6DMG
 
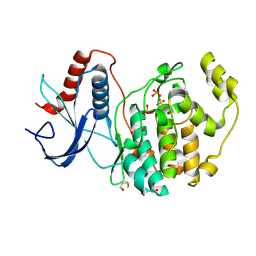 | | A multiconformer ligand model of EK6 bound to ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Hudson, B.M, van Zundert, G.C.P, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMI
 
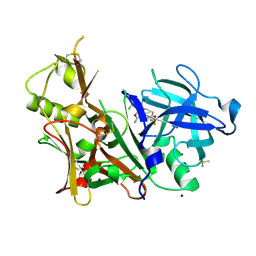 | | A multiconformer ligand model of 5T5 bound to BACE-1 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMK
 
 | | A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMH
 
 | | A multiconformer ligand model of acylenzyme intermediate of meropenem bound to an SFC-1 E166A mutant | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6V0Q
 
 | | Crystal structure of the bromodomain of human BRD7 bound to TG003 | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V0S
 
 | |
6V0X
 
 | | Crystal structure of the bromodomain of human BRD9 bound to sunitinib | | Descriptor: | Bromodomain-containing protein 9, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Karim, M.R, Chan, A, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V16
 
 | | Crystal structure of the bromodomain of human BRD7 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1H
 
 | | Crystal structure of the bromodomain of human BRD7 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1B
 
 | | Crystal structure of the bromodomain of human BRD9 bound to I-BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V14
 
 | | Crystal structure of the bromodomain of human BRD9 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 9 | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V17
 
 | | Crystal structure of the bromodomain of human BRD7 bound to I-BRD9 | | Descriptor: | Bromodomain-containing protein 7, CHLORIDE ION, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6UZF
 
 | | Crystal structure of the unliganded bromodomain of human BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1F
 
 | | Crystal structure of the bromodomain of human BRD7 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
5OR8
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 1,3-dimethyl-benzimidazolone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(4-fluorophenyl)methyl]-1,3,6-trimethyl-2-oxidanylidene-benzimidazole-5-sulfonamide | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5ORB
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methoxyphenyl)sulfanyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5OR9
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5A82
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 4-(3R,4R) -4-(3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)aminopiperidin-3- yloxymethyl)-1-thiane-1,1-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(3R,4R)-3-[[1,1-bis(oxidanylidene)thian-4-yl]methoxy]piperidin-4-yl]amino]-3-methyl-1H-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Optimization of Naphthyridones Into Potent Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5A5Q
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 3-methyl- 8-piperidin-4-ylamino-1,2-dihydro-1,7-naphthyridin-2-one hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-8-[(piperidin-4-yl)amino]-1,2-dihydro-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
7ENZ
 
 | | Crystal structure of Phenanthredinone moiety in complex with the second bromodomain of BRD2 (BRD2-BD2). | | Descriptor: | Bromodomain-containing protein 2, TRIETHYLENE GLYCOL, phenanthridin-6(5H)-one | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ENV
 
 | | crystal structure of NS5 in complex with the N-terminal bromodomain of BRD2 (BRD2-BD1). | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7EO5
 
 | | Crystal structure of pyrano 1,3, oxazine derivative in complex with the second bromodomain of BRD2 | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
