3LW1
 
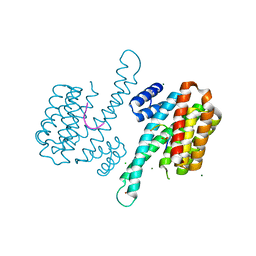 | | Binary complex of 14-3-3 sigma and p53 pT387-peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Schumacher, B, Mondry, J, Thiel, P, Weyand, M, Ottmann, C. | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure of the p53 C-terminus bound to 14-3-3: Implications for stabilization of the p53 tetramer
Febs Lett., 584, 2010
|
|
1JT3
 
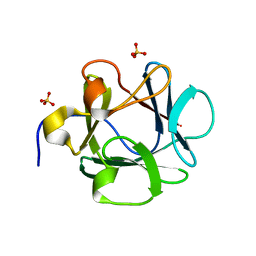 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Histidine Tag AND LEU 73 REPLACED BY VAL (L73V) | | Descriptor: | SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
3LUE
 
 | | Model of alpha-actinin CH1 bound to F-actin | | Descriptor: | Actin, cytoplasmic 1, Alpha-actinin-3 | | Authors: | Galkin, V.E, Orlova, A, Salmazo, A, Djinovic-Carugo, K, Egelman, E.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Opening of tandem calponin homology domains regulates their affinity for F-actin.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CD9
 
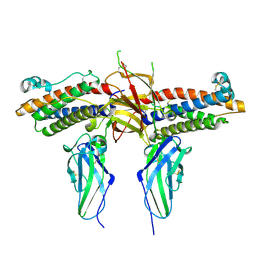 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1H28
 
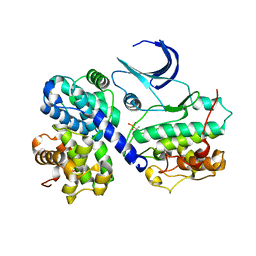 | | CDK2/CyclinA in complex with an 11-residue recruitment peptide from p107 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, RETINOBLASTOMA-LIKE PROTEIN 1 | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
3MS6
 
 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
1I6C
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-02 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1E1X
 
 | |
3MSH
 
 | |
1JSU
 
 | | P27(KIP1)/CYCLIN A/CDK2 COMPLEX | | Descriptor: | CYCLIN A, CYCLIN-DEPENDENT KINASE-2, P27, ... | | Authors: | Russo, A.A, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1996-07-03 | | Release date: | 1997-07-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex.
Nature, 382, 1996
|
|
1H1R
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6086 | | Descriptor: | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor.
Nat. Struct. Biol., 9, 2002
|
|
1H27
 
 | | CDK2/CyclinA in complex with an 11-residue recruitment peptide from p27 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, CYCLIN-DEPENDENT KINASE INHIBITOR 1B | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
1CLD
 
 | | DNA-binding protein | | Descriptor: | CADMIUM ION, CD2-LAC9 | | Authors: | Gardner, K.H, Coleman, J.E. | | Deposit date: | 1995-06-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain.
Nat.Struct.Biol., 2, 1995
|
|
3KAD
 
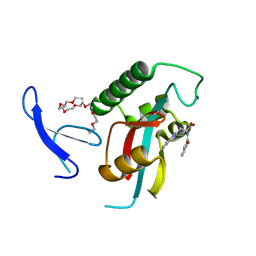 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3JA8
 
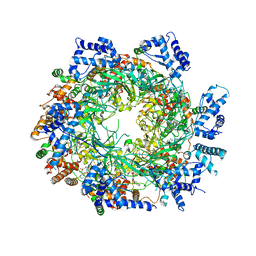 | | Cryo-EM structure of the MCM2-7 double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | Authors: | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
1E4U
 
 | | N-terminal RING finger domain of human NOT-4 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR NOT4, ZINC ION | | Authors: | Hanzawa, H, De Ruwe, M.J, Albert, T.K, Van Der Vliet, P.C, Timmers, H.T, Boelens, R. | | Deposit date: | 2000-07-12 | | Release date: | 2001-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the C4C4 Ring Finger of Human not4 Reveals Features Distinct from Those of C3Hc4 Ring Fingers
J.Biol.Chem., 276, 2001
|
|
3I4B
 
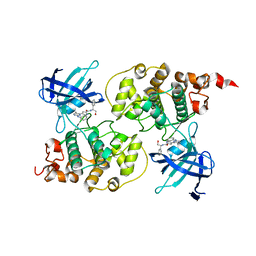 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
1IC8
 
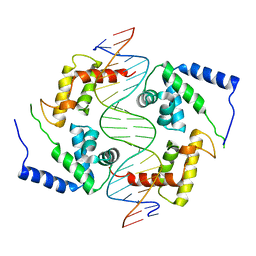 | | HEPATOCYTE NUCLEAR FACTOR 1A BOUND TO DNA : MODY3 GENE PRODUCT | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*G)-3', HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Chi, Y.-I, Frantz, J.D, Oh, B.-C, Hansen, L, Dhe-Paganon, S, Shoelson, S.E. | | Deposit date: | 2001-03-30 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Diabetes mutations delineate an
atypical POU domains in HNF1-Alpha
Mol.Cell, 10, 2002
|
|
1I8H
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH HUMAN TAU PHOSPHOTHREONINE PEPTIDE | | Descriptor: | MICROTUBULE-ASSOCIATED PROTEIN TAU, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1JGG
 
 | | Even-skipped Homeodomain Complexed to AT-rich DNA | | Descriptor: | 5'-D(P*AP*AP*TP*TP*CP*AP*AP*TP*TP*A)-3', 5'-D(P*TP*AP*AP*TP*TP*GP*AP*AP*TP*T)-3', Segmentation Protein Even-Skipped | | Authors: | Hirsch, J.A, Aggarwal, A.K. | | Deposit date: | 2001-06-25 | | Release date: | 2001-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity.
EMBO J., 14, 1995
|
|
3KAG
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-[(2-methylfuran-3-yl)carbonyl]-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3HOM
 
 | |
3KAB
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1F4S
 
 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-09 | | Release date: | 2001-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
1F5E
 
 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-14 | | Release date: | 2001-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
