7ZLM
 
 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound MN551 | | Descriptor: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
8A11
 
 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
5T35
 
 | | The PROTAC MZ1 in complex with the second bromodomain of Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Transcription elongation factor B polypeptide 1, ... | | Authors: | Gadd, M.S, Zengerle, M, Ciulli, A. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of PROTAC cooperative recognition for selective protein degradation.
Nat. Chem. Biol., 13, 2017
|
|
8AQ5
 
 | | KRAS G12C IN COMPLEX WITH GDP AND COMPOUND 16 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-phenyl-pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-08-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | JDQ443, a Structurally Novel, Pyrazole-Based, Covalent Inhibitor of KRAS G12C for the Treatment of Solid Tumors.
J.Med.Chem., 65, 2022
|
|
8AQ7
 
 | | KRAS G12C IN COMPLEX WITH GDP AND COMPOUND 9 | | Descriptor: | 1-[6-[3-cyclohexyl-5-methyl-4-(5-methyl-1~{H}-indazol-4-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-08-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | JDQ443, a Structurally Novel, Pyrazole-Based, Covalent Inhibitor of KRAS G12C for the Treatment of Solid Tumors.
J.Med.Chem., 65, 2022
|
|
5UGI
 
 | |
6TV7
 
 | | Crystal structure of rsGCaMP in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, SODIUM ION, rsGCaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-01-09 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
8BJ4
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in apo form | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Rutkiewicz, M, Witek, W, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ1
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the open state | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ2
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the closed state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ3
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in complex with histidinol-phosphate | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, [(2~{S})-3-(1~{H}-imidazol-4-yl)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]propyl] dihydrogen phosphate, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
7ZU1
 
 | | Crystal structure of mutant AR-LBD (V758A) bound to dihydrotestosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZTV
 
 | | Crystal structure of mutant AR-LBD (F755L) bound to dihydrotestosterone | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
8VL8
 
 | |
7KL7
 
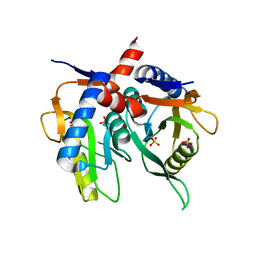 | |
7KL6
 
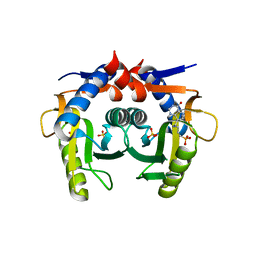 | | Helicobacter pylori Xanthine-Guanine-Hypoxanthine Phosphoribosyltransferase | | Descriptor: | (3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl)phosphonic acid, Phosphoribosyltransferase, SULFATE ION | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Helicobacter pylori Xanthine-Guanine-Hypoxanthine Phosphoribosyltransferase-A Putative Target for Drug Discovery against Gastrointestinal Tract Infections.
J.Med.Chem., 64, 2021
|
|
1OF2
 
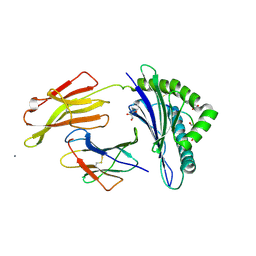 | | Crystal structure of HLA-B*2709 complexed with the vasoactive intestinal peptide type 1 receptor (VIPR) peptide (residues 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HUMAN LYMPHOCYTE ANTIGEN HLA-B27, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-04-04 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual, Hla-B27 Subtype-Dependent Conformation of a Self-Peptide
J.Exp.Med., 199, 2004
|
|
3GTU
 
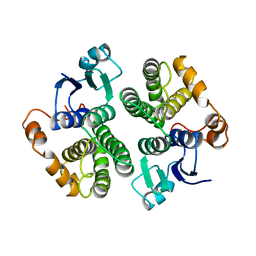 | |
3B6S
 
 | | Crystal Structure of hla-b*2705 Complexed with the Citrullinated Vasoactive Intestinal Peptide Type 1 Receptor (vipr) Peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Citrullination-dependent Differential Presentation of a Self-peptide by HLA-B27 Subtypes.
J.Biol.Chem., 283, 2008
|
|
3BIF
 
 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE EMPTY 6-PF-2K ACTIVE SITE | | Descriptor: | PHOSPHATE ION, PROTEIN (6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE), SUCCINIC ACID, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
Biochemistry, 38, 1999
|
|
2XQN
 
 | | Complex of the 2nd and 3rd LIM domains of TES with the EVH1 DOMAIN of MENA and the N-Terminal domain of actin-like protein Arp7A | | Descriptor: | ACTIN-LIKE PROTEIN 7A, ENABLED HOMOLOG, TESTIN, ... | | Authors: | Knowles, P.P, Briggs, D.C, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2010-09-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
J. Biol. Chem., 286, 2011
|
|
6FL5
 
 | | Structure of human SHMT1-H135N-R137A-E168N mutant at 3.6 Ang. resolution | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Giardina, G, Cutruzzola, F, Lucchi, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells.
FEBS J., 285, 2018
|
|
6FLL
 
 | |
2CBM
 
 | | Crystal structure of the apo-form of a neocarzinostatin mutant evolved to bind testosterone. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NEOCARZINOSTATIN | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
3DTX
 
 | | Crystal structure of HLA-B*2705 complexed with the double citrullinated vasoactive intestinal peptide type 1 receptor (VIPR) peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, Double citrullinated vasoactive intestinal polypeptide receptor, MHC class I antigen (Fragment) | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2008-07-16 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Citrullination-and mhc polymorphism-dependent conformational changes of a self peptide
To be Published
|
|
