4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
8DS8
 
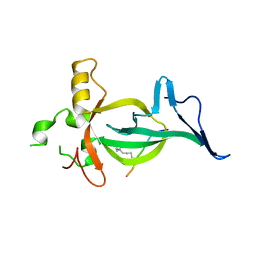 | |
6DZT
 
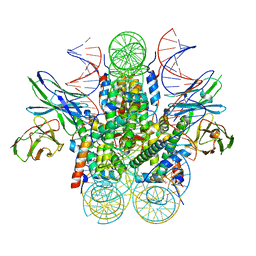 | |
2HIO
 
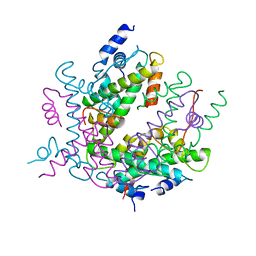 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN | | Descriptor: | PROTEIN (HISTONE H2A), PROTEIN (HISTONE H2B), PROTEIN (HISTONE H3), ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1999-06-15 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6Z2A
 
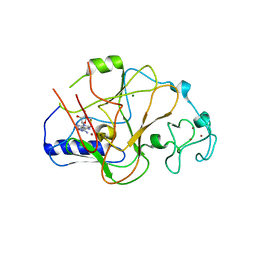 | |
3KXB
 
 | |
5OMX
 
 | |
4FT4
 
 | |
5ONW
 
 | | X-Ray crystal structure of a nucleosome core particle with its DNA site-specifically crosslinked to the histone octamer and the two H2A/H2B dimers crosslinked via H2A N38C | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Frouws, T.D, Barth, P.D, Richmond, T.J. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Site-Specific Disulfide Crosslinked Nucleosomes with Enhanced Stability.
J. Mol. Biol., 430, 2018
|
|
2WP1
 
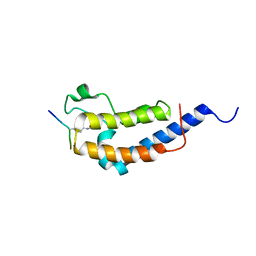 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
7X6L
 
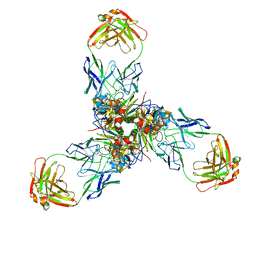 | |
4JJN
 
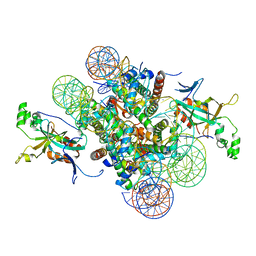 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MUP
 
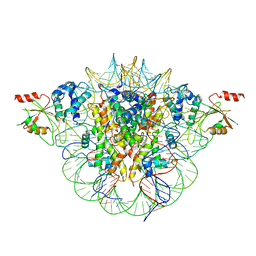 | |
3B95
 
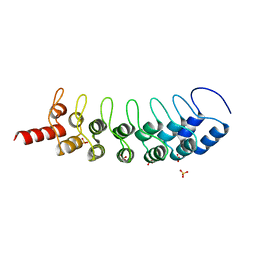 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 2) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, Histone H3 N-terminal Peptide, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6R1T
 
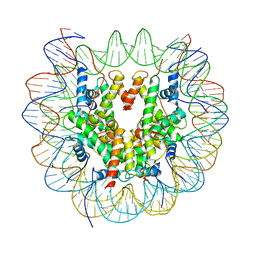 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6MUO
 
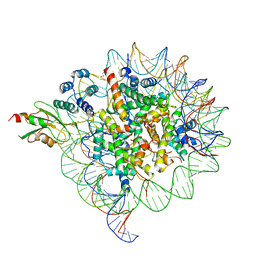 | |
7D20
 
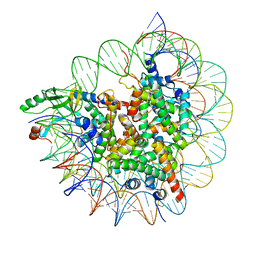 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
6TEM
 
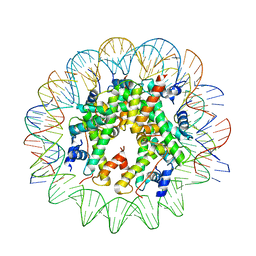 | | CENP-A nucleosome core particle with 145 base pairs of the Widom 601 sequence by cryo-EM | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3-like centromeric protein A, ... | | Authors: | Boopathi, R, Danev, R, Petosa, C, Bednar, J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends.
Nucleic Acids Res., 48, 2020
|
|
2OQ6
 
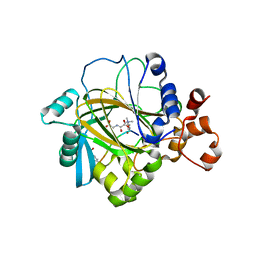 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OS2
 
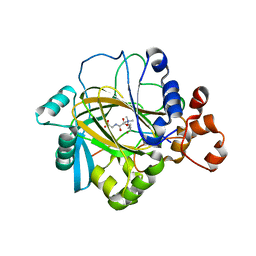 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys36 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2RI7
 
 | |
3NQJ
 
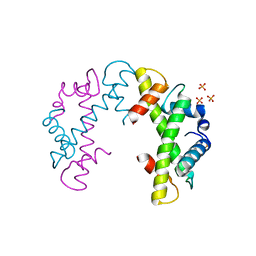 | | Crystal structure of (CENP-A/H4)2 heterotetramer | | Descriptor: | Histone H3-like centromeric protein A, Histone H4, PHOSPHATE ION | | Authors: | Sekulic, N, Black, B.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres.
Nature, 467, 2010
|
|
2OX0
 
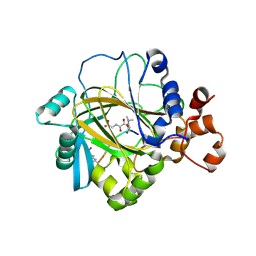 | | Crystal structure of JMJD2A complexed with histone H3 peptide dimethylated at Lys9 | | Descriptor: | CHLORIDE ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
5JJO
 
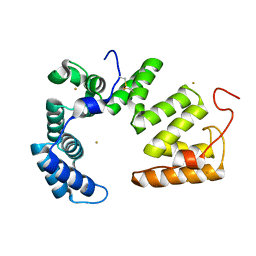 | | The crystal structure of immunity protein PA5088 from Pseudomonas aeruginosa | | Descriptor: | GOLD ION, Uncharacterized protein | | Authors: | Li, Z.Q, Gao, Z.Q, She, Z, Dong, Y.H. | | Deposit date: | 2016-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural analysis of Pseudomonas aeruginosa H3-T6SS immunity proteins
Febs Lett., 590, 2016
|
|
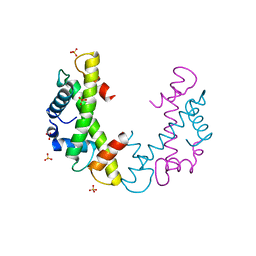
3NQU
 
 | |
