5VNZ
 
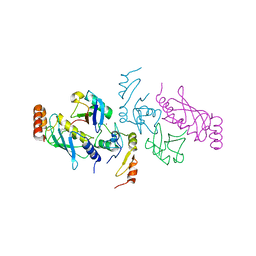 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
5VO0
 
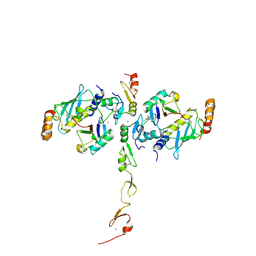 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | POTASSIUM ION, TNF receptor-associated factor 6, Ubiquitin, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
6V0V
 
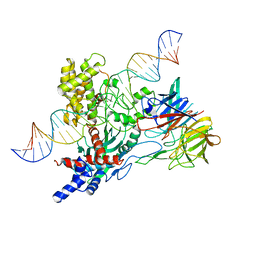 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5H7S
 
 | |
5TTE
 
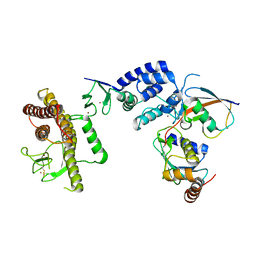 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
8GCY
 
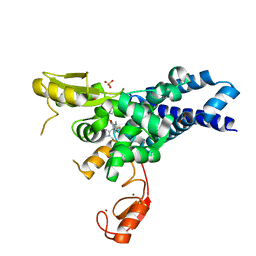 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
7B5N
 
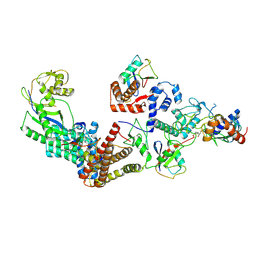 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5S
 
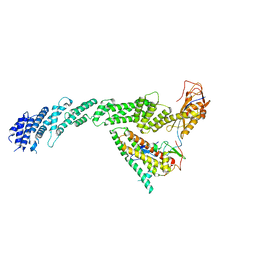 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5M
 
 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
3JBX
 
 | | Cryo-electron microscopy structure of RAG Signal End Complex (C2 symmetry) | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', 5'-D(*GP*CP*GP*AP*TP*GP*GP*TP*TP*AP*AP*CP*CP*A)-3', 5'-D(P*GP*TP*CP*TP*GP*TP*AP*GP*CP*AP*CP*TP*GP*TP*G)-3', ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3JBW
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
8GRQ
 
 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GUJ
 
 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUI
 
 | | Bre1-nucleosome complex (Model I) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
3JBY
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (C2 symmetry) | | Descriptor: | '-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', 5'-D(P*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3KNV
 
 | |
6DBU
 
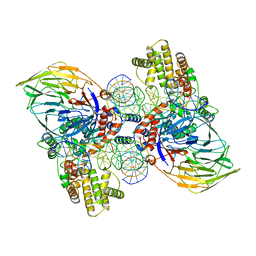 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
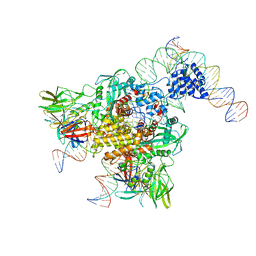 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3VGO
 
 | |
6FGA
 
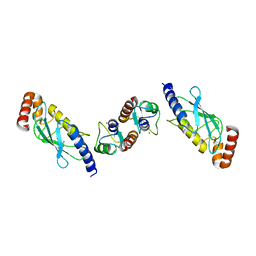 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
4A4C
 
 | | Structure of phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A4B
 
 | | Structure of modified phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A49
 
 | | Structure of phosphoTyr371-c-Cbl-UbcH5B complex | | Descriptor: | E3 ubiquitin-protein ligase CBL, POTASSIUM ION, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl.
Nat. Struct. Mol. Biol., 19, 2012
|
|
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBJ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
