4UD8
 
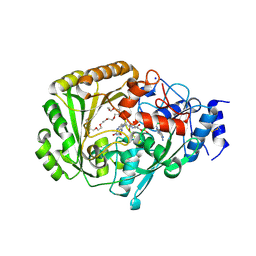 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
1OAV
 
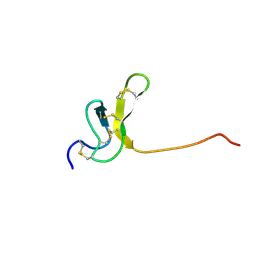 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
7T4E
 
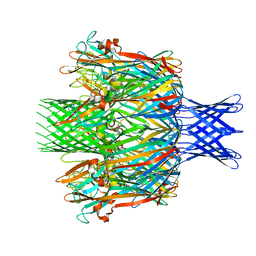 | |
7T4D
 
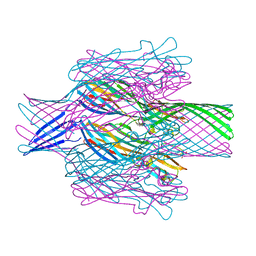 | | Pore structure of pore-forming toxin Epx4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | Authors: | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
4UEX
 
 | |
4LW0
 
 | | Structure of the THF riboswitch bound to adenine | | Descriptor: | ADENINE, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LYP
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SOG
 
 | | Thermostable actophorin | | Descriptor: | Actophorin, NICKEL (II) ION | | Authors: | Lieberman, R.L, Quirk, S. | | Deposit date: | 2021-10-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and activity of a thermally stable mutant of Acanthamoeba actophorin.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7T8V
 
 | | Co-crystal structure of Chaetomium glucosidase I with EB-0159 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T66
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-4 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T68
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-5 | | Descriptor: | (2R,3R,4R,5S)-1-[6-(4-azido-2-nitroanilino)hexyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T6W
 
 | | Crystal structure of Chaetomium Glucosidase I (apo) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, GLYCEROL, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
4USC
 
 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
4MS6
 
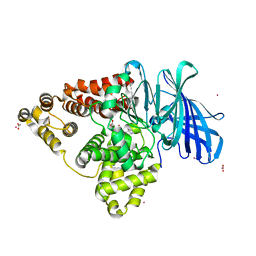 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, ACETIC ACID, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UQM
 
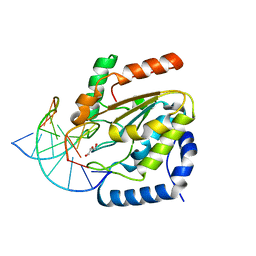 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4UU0
 
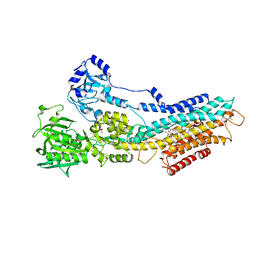 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF 14:1 PC | | Descriptor: | GLYCEROL, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
4WD4
 
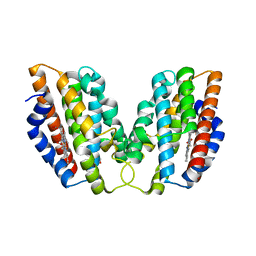 | | Crystal structure of human HO1 H25R | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
4V0Z
 
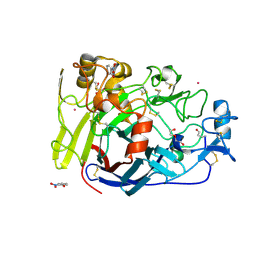 | |
4LFT
 
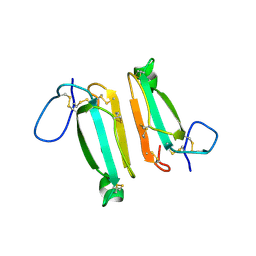 | | Structure of alpha-elapitoxin-Dpp2d isolated from Black Mamba (Dendroaspis polylepis) venom | | Descriptor: | Alpha-elapitoxin-Dpp2a | | Authors: | Wang, C.I.A, Reeks, T, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2013-06-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Isolation and Structural and Pharmacological Characterization of alpha-Elapitoxin-Dpp2d, an Amidated Three Finger Toxin from Black Mamba Venom.
Biochemistry, 53, 2014
|
|
5KBS
 
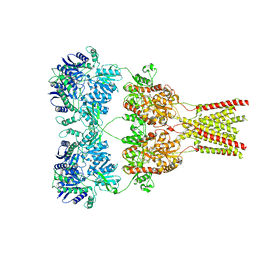 | | Cryo-EM structure of GluA2-0xSTZ at 8.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBU
 
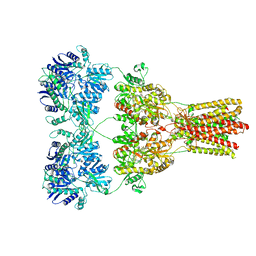 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
4L2L
 
 | | Human Leukotriene A4 Hydrolase complexed with ligand 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-06-04 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJC
 
 | |
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
