3W1G
 
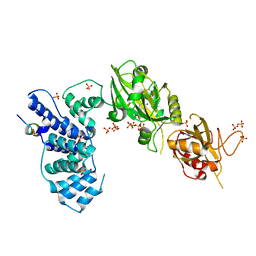 | | Crystal Structure of Human DNA ligase IV-Artemis Complex (Native) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Artemis-derived peptide, DNA ligase 4, ... | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
5TT6
 
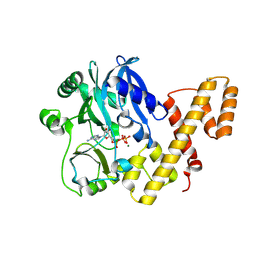 | | T4 RNA Ligase 1 (K99M) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, T4 RNA ligase 1 | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7ELT
 
 | |
7EL8
 
 | |
5C7M
 
 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
7ENS
 
 | |
7ENT
 
 | |
3SY2
 
 | |
2RNN
 
 | |
5TT5
 
 | | Escherichia coli LigA (K115M) in complex with NAD+ | | Descriptor: | DNA ligase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3W1B
 
 | |
3W5O
 
 | | Crystal Structure of Human DNA ligase IV | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA ligase 4, SULFATE ION | | Authors: | Gu, X, Ochi, T, Blundell, T.L. | | Deposit date: | 2013-02-02 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
8IUG
 
 | | Cryo-EM structure of the RC-LH core complex from roseiflexus castenholzii | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-03-24 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
6U1C
 
 | |
8IUN
 
 | | Cryo-EM structure of the CRT-LESS RC-LH core complex from roseiflexus castenholzii | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-03-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
6U1D
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6U1E
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6JOM
 
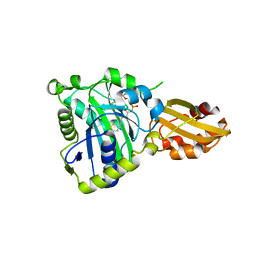 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
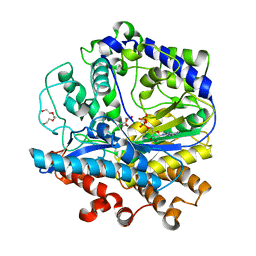
3IG8
 
 | |
4DQ2
 
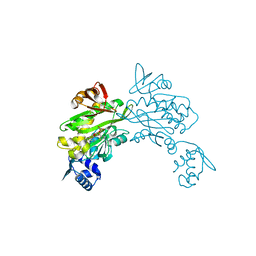 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
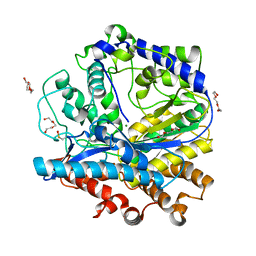
3IG5
 
 | |
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
4ME6
 
 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
7MO6
 
 | |
3PC7
 
 | |
