1AHN
 
 | | E. COLI FLAVODOXIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1997-04-07 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A flavodoxin that is required for enzyme activation: the structure of oxidized flavodoxin from Escherichia coli at 1.8 A resolution.
Protein Sci., 6, 1997
|
|
2NQG
 
 | | Calpain 1 proteolytic core inactivated by WR18(S,S), an epoxysuccinyl-type inhibitor. | | Descriptor: | 5-AZANYLIDYNE-N-[(2S)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]-L-NORVALYL-L-ARGINYL-L-TRYPTOPHANAMIDE, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | Authors: | Yamasaki, M, Takahashi, N, Hirose, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
7JGZ
 
 | | Protocadherin gammaC4 EC1-4 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
3ECG
 
 | | High Resolution HIV-2 Protease Structure in Complex with Antiviral Inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
3EDD
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3QKP
 
 | | Protein Tyrosine Phosphatase 1B - Apo W179F mutant with open WPD-loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2011-02-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B.
Arch.Biochem.Biophys., 525, 2012
|
|
4CKU
 
 | | Three dimensional structure of plasmepsin II in complex with hydroxyethylamine-based inhibitor | | Descriptor: | 5-[1,1-bis(oxidanylidene)-1,2-thiazinan-2-yl]-N3-[(2S,3R)-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-butan-2-yl]-N1,N1-dipropyl-benzene-1,3-dicarboxamide, PLASMEPSIN-2 | | Authors: | Tars, K, Leitans, J, Jaudzems, K. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasmepsin Inhibitory Activity and Structure-Guided Optimization of a Potent Hydroxyethylamine-Based Antimalarial Hit.
Acs Med.Chem.Lett., 5, 2014
|
|
5XPO
 
 | | Crystal structure of VDR-LBD complexed with 25-(hydroxyphenyl)-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (5~{R})-5-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexyl idene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-(4-hydroxyphenyl)hexan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
2KTS
 
 | | NMR structure of the protein NP_415897.1 | | Descriptor: | Heat shock protein hslJ | | Authors: | Serrano, P, Jaudzems, K, Geralt, M, Horst, R, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_415897.1
To be Published
|
|
4OJE
 
 | |
4O8P
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to xylotetraose | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
5XPL
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3VKF
 
 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1J2Z
 
 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
5QCO
 
 | | Crystal structure of BACE complex with BMC016 | | Descriptor: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
2DQU
 
 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
3RS4
 
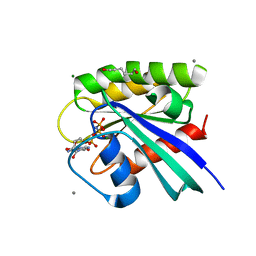 | | H-Ras soaked in 60% 1,6-hexanediol: 1 of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, HEXANE-1,6-DIOL, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
2JIU
 
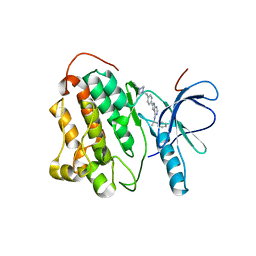 | | Crystal structure of EGFR kinase domain T790M mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JHM
 
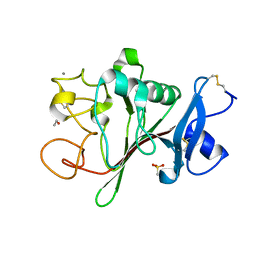 | | Structure of globular heads of M-ficolin at neutral pH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FICOLIN-1, ... | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
4LK0
 
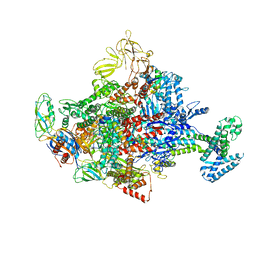 | | Crystal Structure Analysis of the E.coli holoenzyme/T7 Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.9103 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JEM
 
 | | Native family 12 xyloglucanase from Bacillus licheniformis | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K.S, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2OF0
 
 | | X-ray crystal structure of beta secretase complexed with compound 5 | | Descriptor: | (2S)-1-(2,5-dimethylphenoxy)-3-morpholin-4-ylpropan-2-ol, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-02 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to beta-Secretase.
J.Med.Chem., 50, 2007
|
|
