2K4A
 
 | |
1SSY
 
 | | Crystal structure of phage T4 lysozyme mutant G28A/I29A/G30A/C54T/C97A | | Descriptor: | Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KIR
 
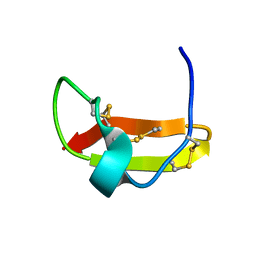 | | Solution structure of a designer toxin, mokatoxin-1 | | Descriptor: | Designer toxin | | Authors: | Biancalana, M, Koide, A, Takacs, Z, Goldstein, S, Koide, S. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1R31
 
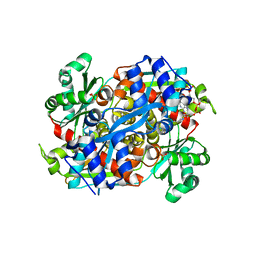 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
1WUF
 
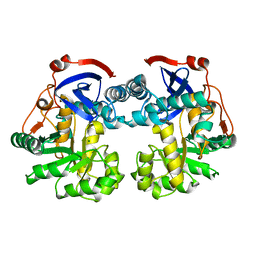 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2M52
 
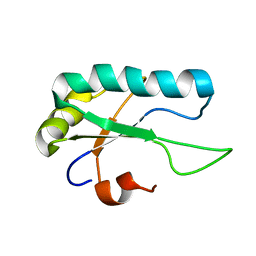 | |
1PL9
 
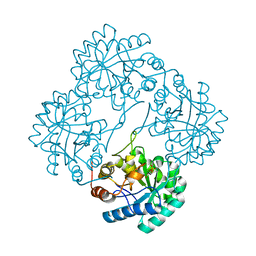 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog Z-FPEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, 3-FLUORO-2-(PHOSPHONOOXY)PROPANOIC ACID | | Authors: | Vainer, R, Adir, N, Baasov, T, Belakhov, V, Rabkin, E. | | Deposit date: | 2003-06-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of KDO8P synthase in its binary complex with substrate analog Z-FPEP
To be Published
|
|
1EV7
 
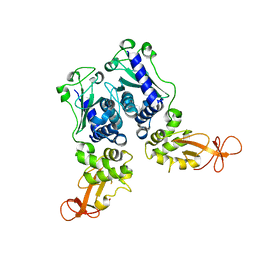 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
2IUL
 
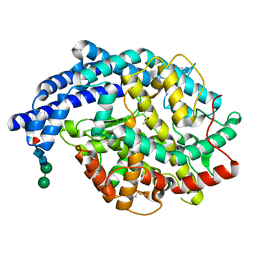 | | Human tACE g13 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Sewell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-06 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
1YRS
 
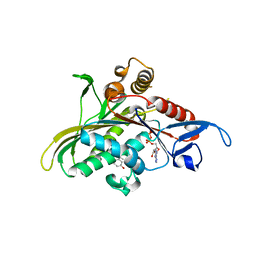 | | Crystal structure of KSP in complex with inhibitor 1 | | Descriptor: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
2KDE
 
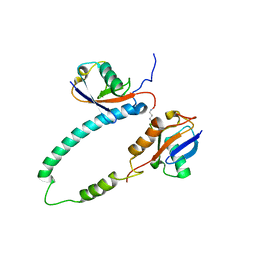 | | NMR structure of major S5a (196-306):K48 linked diubiquitin species | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | Authors: | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
1U0M
 
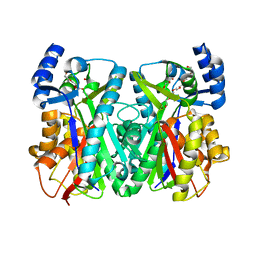 | | Crystal Structure of 1,3,6,8-Tetrahydroxynaphthalene Synthase (THNS) from Streptomyces coelicolor A3(2): a Bacterial Type III Polyketide Synthase (PKS) Provides Insights into Enzymatic Control of Reactive Polyketide Intermediates | | Descriptor: | GLYCEROL, POLYETHYLENE GLYCOL (N=34), putative polyketide synthase | | Authors: | Austin, M.B, Izumikawa, M, Bowman, M.E, Udwary, D.W, Ferrer, J.L, Moore, B.S, Noel, J.P. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates
J.Biol.Chem., 279, 2004
|
|
2A6J
 
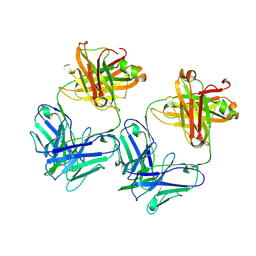 | | Crystal structure analysis of the anti-arsonate germline antibody 36-65 | | Descriptor: | Germline antibody 36-65 Fab heavy chain, Germline antibody 36-65 Fab light chain | | Authors: | Sethi, D.K, Agarwal, A, Manivel, V, Rao, K.V, Salunke, D.M. | | Deposit date: | 2005-07-02 | | Release date: | 2006-06-13 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response.
Immunity, 24, 2006
|
|
1H52
 
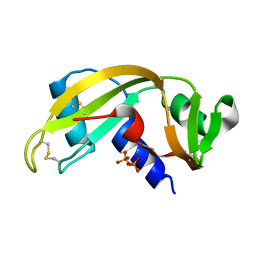 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PYROPHOSPHATE 2- | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1H53
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, CITRIC ACID, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1TYN
 
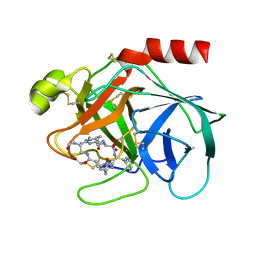 | | ATOMIC STRUCTURE OF THE TRYPSIN-CYCLOTHEONAMIDE A COMPLEX: LESSONS FOR THE DESIGN OF SERINE PROTEASE INHIBITORS | | Descriptor: | BETA-TRYPSIN, CYCLOTHEONAMIDE A | | Authors: | Lee, A.Y, Hagihara, M, Karmacharya, R, Albers, M.W, Schreiber, S.L, Clardy, J. | | Deposit date: | 1994-09-19 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Structure of the Trypsin-Cyclotheonamide a Complex: Lessons for the Design of Serine Protease Inhibitors
J.Am.Chem.Soc., 115, 1993
|
|
2IUX
 
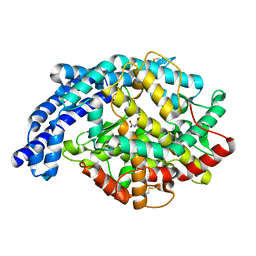 | | Human tACE mutant g1234 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Swell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
1HBY
 
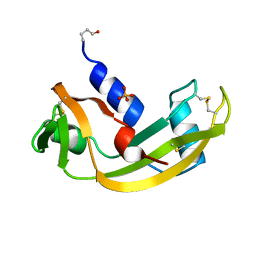 | | Binding of Phosphate and Pyrophosphate ions at the active site of human angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.S, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-04-21 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1Y0Y
 
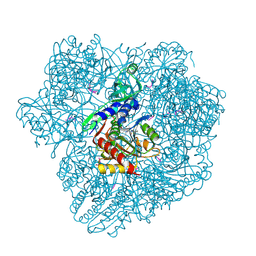 | |
1W77
 
 | | 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) from Arabidopsis thaliana | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CADMIUM ION, COPPER (II) ION, ... | | Authors: | Gabrielsen, M, Kaiser, J, Rohdich, F, Eisenreich, W, Bacher, A, Bond, C.S, Hunter, W.N. | | Deposit date: | 2004-08-30 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Plant 2C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase Exhibits a Distinct Quaternary Structure Compared to Bacterial Homologues and a Possible Role in Feedback Regulation for Cytidine Monophosphate.
FEBS J., 273, 2006
|
|
1Y0R
 
 | |
2HVN
 
 | | Human Aldose Reductase-zopolrestat complex obtained by cocrystallisation after one day (1day_cocryst) | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-07-29 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expect the unexpected or caveat for drug designers: multiple structure determinations using aldose reductase crystals treated under varying soaking and co-crystallisation conditions.
J.Mol.Biol., 363, 2006
|
|
2HVO
 
 | | Human Aldose Reductase-zopolrestat complex obtained by cocrystallisation (10days_cocryst) | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-07-29 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Expect the unexpected or caveat for drug designers: multiple structure determinations using aldose reductase crystals treated under varying soaking and co-crystallisation conditions.
J.Mol.Biol., 363, 2006
|
|
