4PTV
 
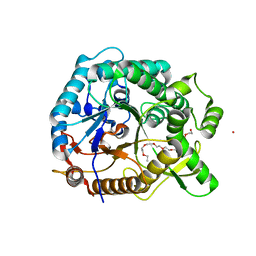 | | Halothermothrix orenii beta-glucosidase A, thiocellobiose complex | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CESIUM ION, Glycoside hydrolase family 1, ... | | Authors: | Hassan, N, Nguyen, T.H, Kori, L.D, Patel, B.K.C, Haltrich, D, Divne, C, Tan, T.C. | | Deposit date: | 2014-03-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
5POC
 
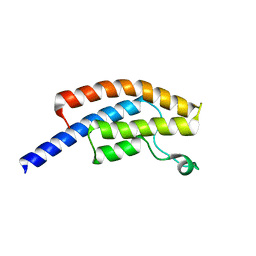 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11081a | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-1-methyl-6-nitroquinolin-2(1H)-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2XOI
 
 | | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-N-[[(2S,3S,4R,5R)-5-(2,4-DIOXOPYRIMIDIN-1-YL)-4-HYDROXY-2-(HYDROXYMETHYL)OXOLAN-3-YL]METHYLSULFONYL]-3,4-DIHYDROXY-OXOLANE-2-CARBOXAMIDE, RIBONUCLEASE PANCREATIC | | Authors: | Thiyagarajan, N, Smith, B.D, Raines, R.T, Acharya, K.R. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A.
FEBS J., 278, 2011
|
|
6B1H
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | Authors: | van den Akker, F, Nhuyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
1G4M
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
6FIC
 
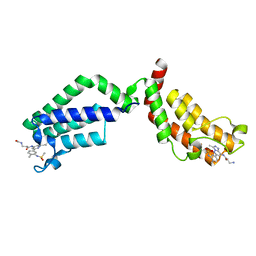 | | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem | | Descriptor: | 3-azanyl-2-[3-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[[3-azanyl-1-[[2-[[3-methyl-6-[4-methyl-3-(methylsulfonyl-$l^{2}-azanyl)cyclohexa-1,3,5-trien-1-yl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]-$l^{2}-azanyl]-2-oxidanylidene-ethyl]amino]-1-oxidanylidene-propan-2-yl]amino]-2-oxidanylidene-ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]-~{N}-[3-[[3-methyl-6-[4-methyl-3-(methylsulfonylamino)phenyl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]-3-oxidanylidene-propyl]propanamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Mathea, S, Suh, J.L, Salah, E, Tallant, C, Siejka, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, James, L.I, Frye, S.V, Knapp, S. | | Deposit date: | 2018-01-17 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem
To Be Published
|
|
1ZFQ
 
 | | carbonic anhydrase II in complex with ethoxzolamidphenole as sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 6-HYDROXY-1,3-BENZOTHIAZOLE-2-SULFONAMIDE, Carbonic anhydrase II, ... | | Authors: | Honndorf, V.S, Heine, A, Klebe, G, Supuran, C.T. | | Deposit date: | 2005-04-20 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | carbonic anhydrase II in complex with ethoxzolamidphenole as sulfonamide inhibitor
To be Published
|
|
2HZN
 
 | | Abl kinase domain in complex with NVP-AFG210 | | Descriptor: | 1-[4-(PYRIDIN-4-YLOXY)PHENYL]-3-[3-(TRIFLUOROMETHYL)PHENYL]UREA, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-09 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4GXH
 
 | |
4OIC
 
 | | Crystal structrual of a soluble protein | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | Authors: | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | Deposit date: | 2014-01-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
4P8W
 
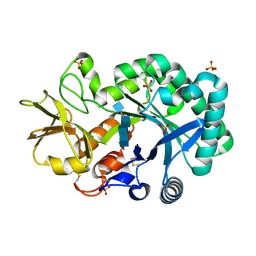 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
5DLT
 
 | | Crystal structure of Autotaxin (ENPP2) with 7-alpha-hydroxycholesterol | | Descriptor: | 7alpha-hydroxycholesterol, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Hausmann, J, Joosten, R.P, Perrakis, A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling.
Nat Commun, 7, 2016
|
|
5T28
 
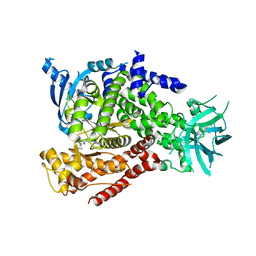 | | mPI3Kd IN COMPLEX WITH 5k | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6AYB
 
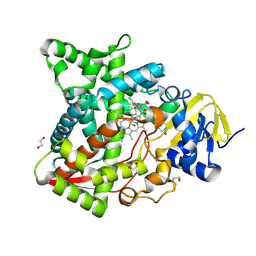 | | Naegleria fowleri CYP51-ketoconazole complex | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, CALCIUM ION, ... | | Authors: | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
4PC7
 
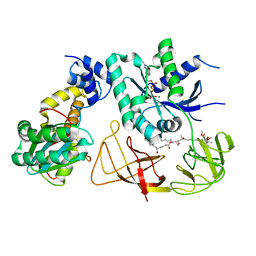 | | Elongation factor Tu:Ts complex in a near GTP conformation. | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6003 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
2ZOT
 
 | |
4OON
 
 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
1R30
 
 | | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Berkovitch, F, Nicolet, Y, Wan, J.T, Jarrett, J.T, Drennan, C.L. | | Deposit date: | 2003-09-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
Science, 303, 2004
|
|
2OGT
 
 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | Descriptor: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
5PNX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10128a | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-1,2-oxazol-3-amine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10919a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-[(4-methoxyphenyl)methyl]acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2OF4
 
 | | crystal structure of furanopyrimidine 1 bound to lck | | Descriptor: | 5,6-DIPHENYL-N-(2-PIPERAZIN-1-YLETHYL)FURO[2,3-D]PYRIMIDIN-4-AMINE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Martin, M.W. | | Deposit date: | 2007-01-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of novel 2,3-diarylfuro[2,3-b]pyridin-4-amines as potent and selective inhibitors of Lck: Synthesis, SAR, and pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OFV
 
 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
5QXJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC578 | | Descriptor: | (3aS,8S,9aS)-10-methyl-4-oxo-1,4,6,8,9,9a-hexahydro-3a,8-epiminocyclohepta[1,2-c:4,5-c']dipyrrole-2(3H)-carbaldehyde, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
1B8E
 
 | |
