8B9G
 
 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
7MSG
 
 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
8BEC
 
 | | Crystal structure of the SARS-CoV-2 S RBD in complex with pT1375 scFV | | Descriptor: | SULFATE ION, Spike protein S1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hansen, G, Ssebyatika, G.L, Krey, T. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
8BG1
 
 | | Crystal structure of the SARS-CoV-2 S RBD in complex with pT1511 scFV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hansen, G, Ssebyatika, G.L, Krey, T. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
8R1X
 
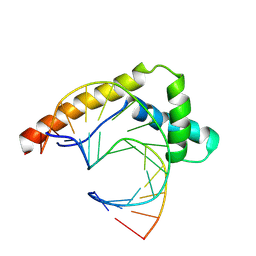 | |
8BG2
 
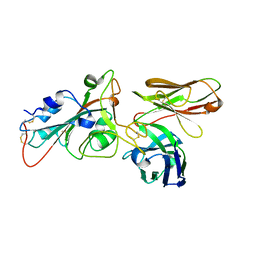 | |
8BG3
 
 | |
7SXN
 
 | | Orb2A residues 1-9 MYNKFVNFI | | Descriptor: | Orb2A residues 1-9 MYNKFVNFI | | Authors: | Bowler, J.T, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2021-11-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like beta-sheets.
J.Biol.Chem., 298, 2022
|
|
8BG5
 
 | |
8BG4
 
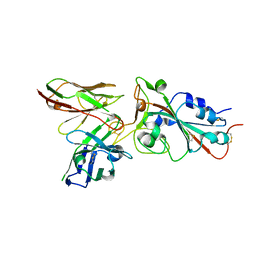 | |
8BG6
 
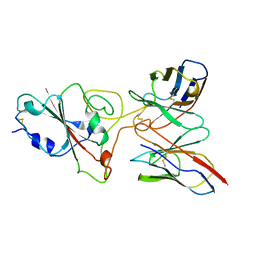 | | SARS-CoV-2 S protein in complex with pT1644 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, pT1644 Fab heavy chain, ... | | Authors: | Stroeh, L, Hansen, G, Vollmer, B, Krey, T, Benecke, T, Gruenewald, K. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
7UL7
 
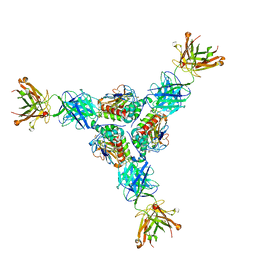 | | Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab | | Descriptor: | 18.5C-M30 Fab Heavy Chain, 18.5C-M30 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Buck, T.K, Enriquez, A.S, Hastie, K.M. | | Deposit date: | 2022-04-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Neutralizing Antibodies against Lassa Virus Lineage I.
Mbio, 13, 2022
|
|
8BOW
 
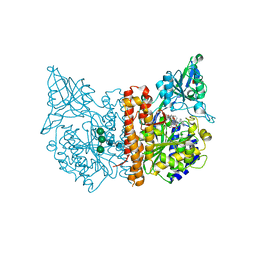 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor 617 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-3-naphthalen-2-yl-2-[[4-[[2-[(11~{Z})-4,7,10-tris(2-hydroxy-2-oxoethyl)-1,4,7,10-tetrazacyclododec-11-en-1-yl]ethanoylamino]methyl]cyclohexyl]carbonylamino]propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Barinka, C, Benesova, M. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor 617
To Be Published
|
|
8BOL
 
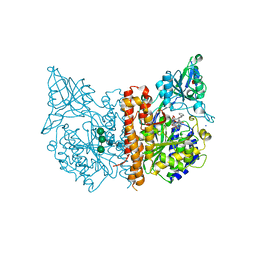 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor P18 | | Descriptor: | (2~{S})-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-6-[[(~{E},2~{S})-5-phenyl-2-[[4-[[2-[4,7,10-tris(2-hydroxy-2-oxoethyl)-1,4,7,10-tetrazacyclododec-1-yl]ethanoylamino]methyl]cyclohexyl]carbonylamino]pent-4-enoyl]amino]hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Barinka, C, Benesova, M. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor P18
To Be Published
|
|
7N7B
 
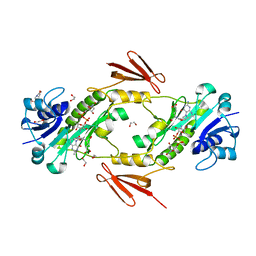 | | crystal structure of the N-formyltrasferase HCAN_0200 from Helicobacter canadensis on complex with folinic acid and dTDP-3-aminofucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
8BO8
 
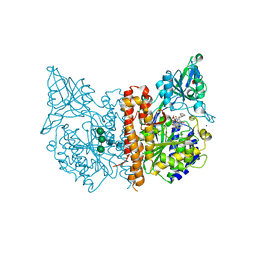 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor P17 | | Descriptor: | (2~{S})-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-6-[[(~{E},2~{S})-5-phenyl-2-[[4-[[2-[4,7,10-tris(2-hydroxy-2-oxoethyl)-1,4,7,10-tetrazacyclododec-1-yl]ethanoylamino]methyl]cyclohexyl]carbonylamino]pent-4-enoyl]amino]hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Barinka, C, Benesova, M. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor P17
To Be Published
|
|
8BW8
 
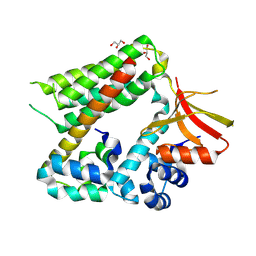 | |
8BW9
 
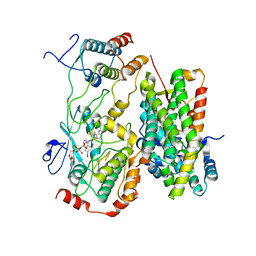 | | Cryo-EM structure of the RAF activating complex KSR-MEK-CNK-HYP | | Descriptor: | Connector enhancer of KSR protein CNK, Dual specificity mitogen-activated protein kinase kinase dSOR1, KSR, ... | | Authors: | Maisonneuve, P, Fronzes, R, Sicheri, F. | | Deposit date: | 2022-12-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The CNK-HYP scaffolding complex promotes RAF activation by enhancing KSR-MEK interaction.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PJJ
 
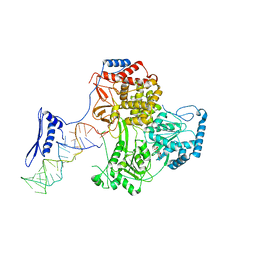 | |
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7JK6
 
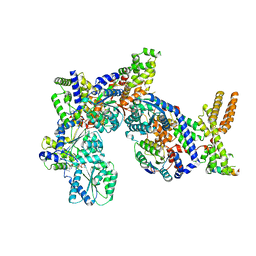 | | Structure of Drosophila ORC in the active conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK2
 
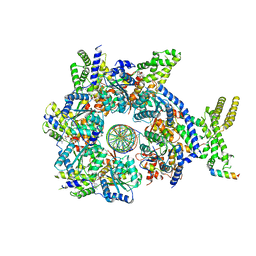 | |
7JGR
 
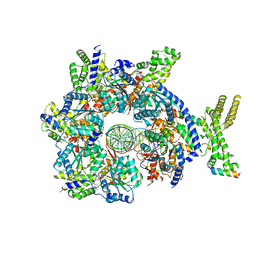 | | Structure of Drosophila ORC bound to DNA (84 bp) and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AT22044p1, Cell division control protein, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK3
 
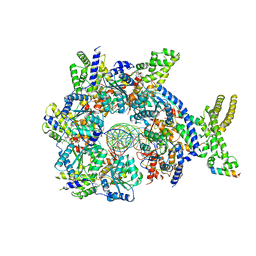 | | Structure of Drosophila ORC bound to GC-rich DNA and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein, DNA (33-MER), ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JK5
 
 | | Structure of Drosophila ORC bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (32-MER), MAGNESIUM ION, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
