2QTN
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Sershon, V.C, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and X-ray structural evidence for negative cooperativity in substrate binding to nicotinate mononucleotide adenylyltransferase (NMAT) from Bacillus anthracis.
J.Mol.Biol., 385, 2009
|
|
3U8W
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Triazolopyridazinone inhibitor | | Descriptor: | 3-[3-(2-chloro-6-fluorophenyl)-5-ethyl-6-oxo-5,6-dihydro[1,2,4]triazolo[4,3-b]pyridazin-7-yl]-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of triazolopyridazinones as potent p38alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1IKN
 
 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
2R5P
 
 | | Crystal Structure Analysis of HIV-1 Subtype C Protease Complexed with Indinavir | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, ... | | Authors: | Coman, R.M, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Contribution of Naturally Occurring Polymorphisms in Altering the Biochemical and Structural Characteristics of HIV-1 Subtype C Protease
Biochemistry, 47, 2008
|
|
1BWN
 
 | | PH DOMAIN AND BTK MOTIF FROM BRUTON'S TYROSINE KINASE MUTANT E41K IN COMPLEX WITH INS(1,3,4,5)P4 | | Descriptor: | BRUTON'S TYROSINE KINASE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A, Potter, B, Saraste, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
2BLG
 
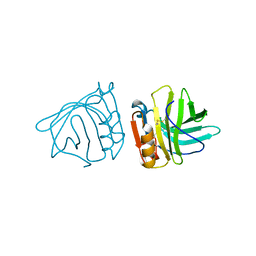 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 8.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
2C29
 
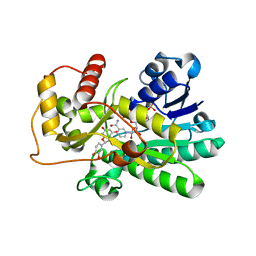 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
2BZW
 
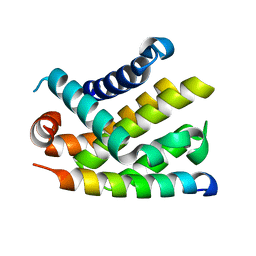 | | The crystal structure of BCL-XL in complex with full-length BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BCL2-ANTAGONIST OF CELL DEATH | | Authors: | Lee, K.-H, Han, W.-D, Kim, K.-J, Oh, B.-H. | | Deposit date: | 2005-08-24 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral Bcl-2 of Murine Gamma-Herpesvirus 68.
Plos Pathog., 4, 2008
|
|
1SJB
 
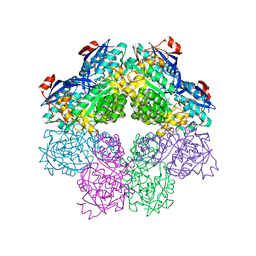 | | X-ray structure of o-succinylbenzoate synthase complexed with o-succinylbenzoic acid | | Descriptor: | 2-SUCCINYLBENZOATE, MAGNESIUM ION, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
4CAT
 
 | | THREE-DIMENSIONAL STRUCTURE OF CATALASE FROM PENICILLIUM VITALE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Melik-Adamyan, W.R, Barynin, V.V, Vagin, A.A, Grebenko, A.I. | | Deposit date: | 1983-02-24 | | Release date: | 1983-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution.
J.Mol.Biol., 188, 1986
|
|
1X9Y
 
 | | The prostaphopain B structure | | Descriptor: | cysteine proteinase | | Authors: | Filipek, R, Szczepanowski, R, Sabat, A, Potempa, J, Bochtler, M. | | Deposit date: | 2004-08-24 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prostaphopain B structure: a comparison of proregion-mediated and staphostatin-mediated protease inhibition.
Biochemistry, 43, 2004
|
|
9BNA
 
 | |
3V89
 
 | | The crystal structure of transferrin binding protein A (TbpA) from Neisseria meningitidis serogroup B in complex with the C-lobe of human transferrin | | Descriptor: | Serotransferrin, Transferrin-binding protein A | | Authors: | Noinaj, N, Oke, M, Easley, N, Zak, O, Aisen, P, Buchanan, S.K. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
3Q8H
 
 | | Crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from burkholderia pseudomallei in complex with cytidine derivative EBSI01028 | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-deoxy-5'-[(imidazo[2,1-b][1,3]thiazol-5-ylcarbonyl)amino]cytidine, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytidine derivatives as IspF inhibitors of Burkolderia pseudomallei.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M20
 
 | |
3E2J
 
 | |
5ENV
 
 | | YEAST ALCOHOL DEHYDROGENASE WITH BOUND COENZYME | | Descriptor: | Alcohol dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Plapp, B.V, Charlier Jr, H.A, Ramaswamy, S. | | Deposit date: | 2015-11-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic implications from structures of yeast alcohol dehydrogenase complexed with coenzyme and an alcohol.
Arch.Biochem.Biophys., 591, 2015
|
|
4M0E
 
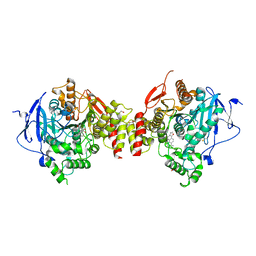 | | Structure of human acetylcholinesterase in complex with dihydrotanshinone I | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Gary, E.N, Shiomi, K, Rosenberry, T.L. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human acetylcholinesterase bound to dihydrotanshinone I and territrem B show peripheral site flexibility.
ACS Med Chem Lett, 4, 2013
|
|
1YN4
 
 | | Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens | | Descriptor: | EapH1, ZINC ION | | Authors: | Geisbrecht, B.V, Hamaoka, B.Y, Perman, B, Zemla, A, Leahy, D.J. | | Deposit date: | 2005-01-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structures of EAP Domains from Staphylococcus aureus Reveal an Unexpected Homology to Bacterial Superantigens.
J.Biol.Chem., 280, 2005
|
|
1F6V
 
 | |
3VT0
 
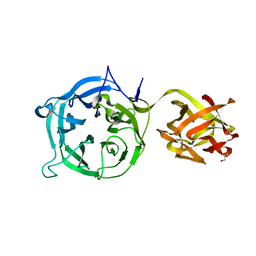 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
4MNB
 
 | | Crystal Structure of a complex between the marine anticancer drug Variolin B and DNA | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, COBALT (II) ION, ... | | Authors: | Canals, A, Arribas-Bosacoma, R, Alvarez, M, Albericio, F, Aymami, J, Coll, M. | | Deposit date: | 2013-09-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Intercalative DNA binding of the marine anticancer drug variolin B.
Sci Rep, 7, 2017
|
|
1YQ9
 
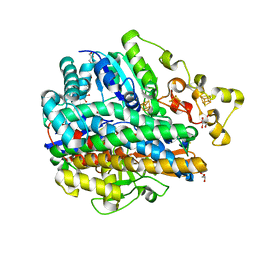 | | Structure of the unready oxidized form of [NiFe] hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
1NXR
 
 | | HIV-1 POLYPURINE HYBRID, R(GAGGACUG):D(CAGTCCTC), NMR, 18 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*TP*C)-3'), RNA (5'-R(*GP*AP*GP*GP*AP*CP*UP*G)-3') | | Authors: | Fedoroff, O.Y, Ge, Y, Reid, B.R. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis.
J.Mol.Biol., 269, 1997
|
|
1LWE
 
 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
