7T2C
 
 | | Crystal structure of the B5 TCR in complex with HLA-DP4-Ply | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ciacchi, L, Farenc, C, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | CD4 + T cell-mediated recognition of a conserved cholesterol-dependent cytolysin epitope generates broad antibacterial immunity.
Immunity, 56, 2023
|
|
4OVY
 
 | | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Chang, C, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776
To be published
|
|
1MZI
 
 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
5SHU
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc2c(c1)CCN2)[nH]c(n3)SCc4ncc(c(c4C)OC)C, micromolar IC50=0.2659405 | | Descriptor: | 2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1,5,6,7-tetrahydroimidazo[4,5-f]indole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Krasso, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7BYM
 
 | | Cryo-EM structure of human KCNQ4 with retigabine | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
5SHG
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(nc2c(cc1)nc(c3c(nc(n23)c4ccccc4Cl)C)N)OC, micromolar IC50=0.001022 | | Descriptor: | (10R)-9-(2-chlorophenyl)-2-methoxy-7-methylimidazo[1,5-a]pyrido[3,2-e]pyrazin-6-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
6K6N
 
 | | Crystal structure of SIVmac239 Nef protein | | Descriptor: | Protein Nef | | Authors: | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.0002 Å) | | Cite: | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
7F5T
 
 | | Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
5ZTY
 
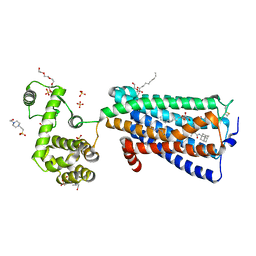 | | Crystal structure of human G protein coupled receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | Deposit date: | 2018-05-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
5SJ8
 
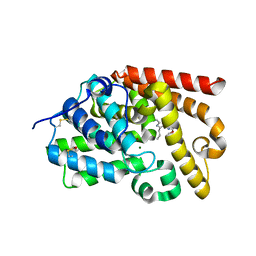 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(OC)nc2c(c1)nc(c3c(nc(n23)CCC)C)C#N, micromolar IC50=0.013985 | | Descriptor: | (10S)-2-methoxy-7-methyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine-6-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SHX
 
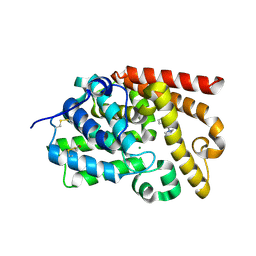 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(nc2c(nc1C)cccc2)N3CCCC3, micromolar IC50=9.069546 | | Descriptor: | 2-methyl-3-(pyrrolidin-1-yl)quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Canesso, R, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7XO7
 
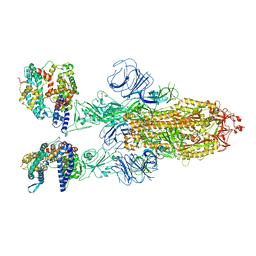 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
3PRI
 
 | | Crystal Structure of Human B-Raf Kinase in Complex with a Non-Oxime Furopyridine Inhibitor | | Descriptor: | 3-(4-{[2-(pyrimidin-2-yl)furo[2,3-c]pyridin-3-yl]amino}-1H-indazol-3-yl)propan-1-ol, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Vigers, G.P.A, Morales, T, Brandhuber, B.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Non-oxime inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6Q8J
 
 | | Nterminal domain of human SMU1 in complex with LSP641 | | Descriptor: | 1-~{tert}-butyl-3-[6-(3,5-dimethoxyphenyl)-2-[[1-[1-(phenylmethyl)piperidin-4-yl]-1,2,3-triazol-4-yl]methylamino]pyrido[2,3-d]pyrimidin-7-yl]urea, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GCC
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
3DR7
 
 | | GDP-perosamine synthase from Caulobacter crescentus with bound GDP-3-deoxyperosamine | | Descriptor: | (2R,3S,5S,6R)-5-amino-3-hydroxy-6-methyl-oxan-2-yl, 1,2-ETHANEDIOL, Putative perosamine synthetase | | Authors: | Holden, H.M, Cook, P.D, Carney, A.E. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of GDP-linked sugars in the active site of GDP-perosamine synthase
Biochemistry, 47, 2008
|
|
8DLW
 
 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with Fab S2M11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S2M11 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
2DSK
 
 | | Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, chitinase | | Authors: | Nakamura, T, Mine, S, Hagihara, Y, Ishikawa, K, Uegaki, K. | | Deposit date: | 2006-06-30 | | Release date: | 2007-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the catalytic domain of the hyperthermophilic chitinase from Pyrococcus furiosus
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
6K9H
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
7BQ1
 
 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin)-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, PALMITIC ACID, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQF
 
 | | Dimerization of SAV1 WW tandem | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Protein salvador homolog 1 | | Authors: | Lin, Z, Zhang, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.70037615 Å) | | Cite: | A WW Tandem-Mediated Dimerization Mode of SAV1 Essential for Hippo Signaling.
Cell Rep, 32, 2020
|
|
6A5W
 
 | | FXR-LBD with HNC143 and SRC1 | | Descriptor: | 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
3GVG
 
 | |
