3GY7
 
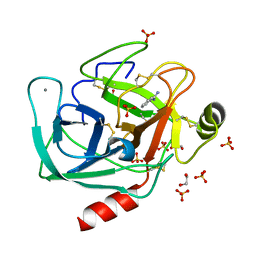 | | A comparative study on the inhibition of bovine beta-trypsin by bis-benzamidines diminazene and pentamidine by X-ray crystallography and ITC | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Perilo, C.S, Pereira, M.T, Santoro, M.M, Nagem, R.A.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural binding evidence of the trypanocidal drugs Berenil and Pentacarinate active principles to a serine protease model.
Int.J.Biol.Macromol., 46, 2010
|
|
3E3T
 
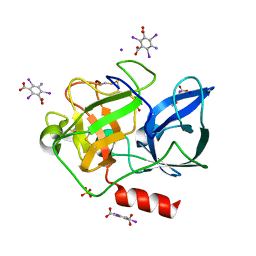 | | Structure of porcine pancreatic elastase with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Elastase-1, IODIDE ION, ... | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-28 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E3S
 
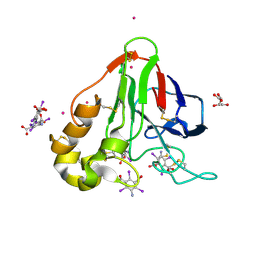 | | Structure of thaumatin with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E64
 
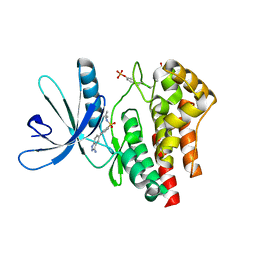 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 4-(3-amino-1H-indazol-5-yl)-N-tert-butylbenzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E3D
 
 | | Structure of hen egg white lysozyme with the magic triangle I3C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Lysozyme C | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-07 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EKO
 
 | |
3EKR
 
 | | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone | | Descriptor: | 4-{[(2R)-2-(2-methylphenyl)pyrrolidin-1-yl]carbonyl}benzene-1,3-diol, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2008-09-19 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6O52
 
 | |
6O50
 
 | | Binary complex of native hAChE with BW284c51 | | Descriptor: | 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O43
 
 | | Crystal structure of a lysin protein from Staphylococcus phage P68 | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Orf11 | | Authors: | Truong, J.Q. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08222985 Å) | | Cite: | Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3D27
 
 | | E. coli methionine aminopeptidase with Fe inhibitor W29 | | Descriptor: | 4-(3-ethylthiophen-2-yl)benzene-1,2-diol, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Chai, S, He, H.Z. | | Deposit date: | 2008-05-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of inhibitors of Escherichia coli methionine aminopeptidase with the Fe(II)-form selectivity and antibacterial activity.
J.Med.Chem., 51, 2008
|
|
3CID
 
 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CJG
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-methyl-N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3CPB
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a bisamide inhibitor | | Descriptor: | N'-(6-aminopyridin-3-yl)-N-(2-cyclopentylethyl)-4-methyl-benzene-1,3-dicarboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Aryl Aminoquinazoline Pyridones as Potent, Selective, and Orally Efficacious Inhibitors of Receptor Tyrosine Kinase c-Kit.
J.Med.Chem., 51, 2008
|
|
3DA2
 
 | | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 13, N-(4-chlorobenzyl)-N-methylbenzene-1,4-disulfonamide, ... | | Authors: | Pilka, E.S, Picaud, S.S, Yue, W.W, King, O.N.F, Bray, J.E, Filippakopoulos, P, Roos, A.K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor.
To be Published
|
|
3DM6
 
 | | Beta-secretase 1 complexed with statine-based inhibitor | | Descriptor: | 5-[[(2S)-2-[[(3R,4S)-5-(3,5-difluorophenoxy)-3-hydroxy-4-[[3-(methyl-methylsulfonyl-amino)-5-[[(1R)-1-phenylethyl]carbamoyl]phenyl]carbonylamino]pentanoyl]amino]-3-methyl-butanoyl]amino]benzene-1,3-dicarboxylic acid, Beta-secretase 1, ISOPROPYL ALCOHOL | | Authors: | Lindberg, J, Borkakoti, N, Nystrom, S. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and SAR of potent statine-based BACE-1 inhibitors: exploration of P1 phenoxy and benzyloxy residues
Bioorg.Med.Chem., 16, 2008
|
|
6PBB
 
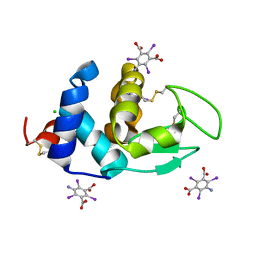 | | Crystal structure of Hen Egg White Lysozyme in complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, Lysozyme | | Authors: | Truong, J.Q. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.89151084 Å) | | Cite: | Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6P12
 
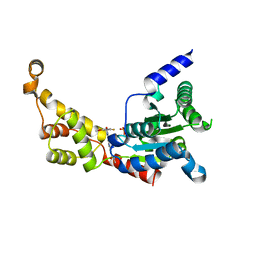 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94131851 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P14
 
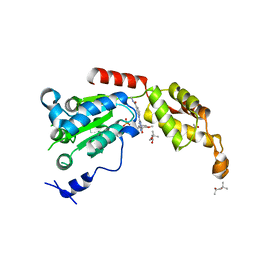 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P13
 
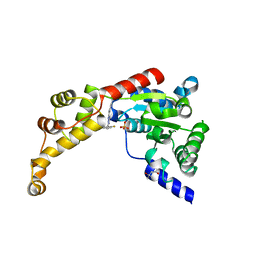 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form A) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P9B
 
 | |
6PZP
 
 | | Crystal structure of caspase-1 in complex with VX-765 | | Descriptor: | Caspase-1, N-(4-amino-3-chlorobenzene-1-carbonyl)-3-methyl-L-valyl-N-[(2S)-1-carboxy-3-oxopropan-2-yl]-L-prolinamide | | Authors: | Yang, J, Liu, Z, Xiao, T.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of caspase-1 in complex with VX-765
To Be Published
|
|
3FTX
 
 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3E33
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 7 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-2-methylbenzenesulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
