2HTJ
 
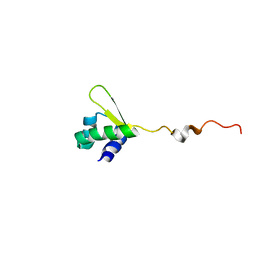 | | NMR structure of E.coli PapI | | Descriptor: | P fimbrial regulatory protein KS71A | | Authors: | Kawamura, T, Zhou, H, Le, L.U.K, Dahlquist, F.W. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Escherichia coli PapI, a Key Regulator of the Pap Pili Phase Variation.
J.Mol.Biol., 365, 2007
|
|
2GSZ
 
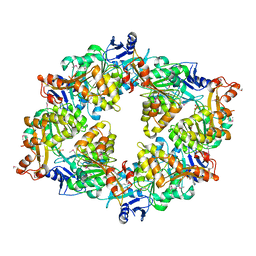 | |
3ZCJ
 
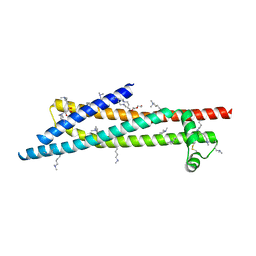 | |
3ZCI
 
 | |
2Y4Y
 
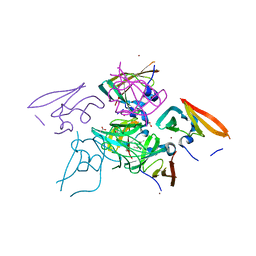 | |
3CNR
 
 | |
6N0A
 
 | | Structure of the major pilin protein (T-18.1) from Streptococcus pyogenes serotype MGAS8232 | | Descriptor: | CALCIUM ION, Major pilin backbone protein T-antigen | | Authors: | Young, P.G, Raynes, J.M, Loh, J.M, Proft, T, Baker, E.N, Moreland, N.J. | | Deposit date: | 2018-11-06 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
4NOA
 
 | |
4XA2
 
 | |
4CII
 
 | | Crystal structure of N-terminally truncated Helicobacter pylori T4SS Protein CagL as domain swapped dimer | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAG PATHOGENICITY ISLAND PROTEIN (CAG18) | | Authors: | Barden, S, Schomburg, B, Niemann, H.H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-19 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a Three-Dimensional Domain-Swapped Dimer of the Helicobacter Pylori Type Iv Secretion System Pilus Protein Cagl.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KYH
 
 | | Structure of Iho670 Flagellar-like Filament | | Descriptor: | Iho670 | | Authors: | Braun, T, Vos, M, Kalisman, N, Sherman, N.E, Rachel, R, Wirth, R, Schroeder, G.F, Egelman, E.H. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2LXQ
 
 | | Monomeric PilE G-Quadruplex DNA from Neisseria Gonorrhoeae | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*T)-3') | | Authors: | Kuryavyi, V.V, Patel, D.J. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RecA-Binding pilE G4 Sequence Essential for Pilin Antigenic Variation Forms Monomeric and 5' End-Stacked Dimeric Parallel G-Quadruplexes.
Structure, 20, 2012
|
|
2LXV
 
 | |
4LW9
 
 | | Crystal structure of Vibrio cholera major pseudopilin EpsG | | Descriptor: | CALCIUM ION, CHLORIDE ION, PLATINUM (II) ION, ... | | Authors: | Vago, F.S, Raghunathan, K, Jens, J.C, Wedemeyer, W.J, Bagdasarian, M, Brunzelle, J.S, Arvidson, D.N. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Vibrio cholera major pseudopilin EpsG
To be Published
|
|
7QAC
 
 | | The T2 structure of polycrystalline cubic human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Karavassili, F, Triandafillidis, D.P, Valmas, A, Spiliopoulou, M, Fili, S, Kontou, P, Bowler, M.W, Von Dreele, R.B, Fitch, A, Margiolaki, I. | | Deposit date: | 2021-11-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | POWDER DIFFRACTION (2.29 Å) | | Cite: | The T 2 structure of polycrystalline cubic human insulin.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6LCH
 
 | | Pseudomonas aeruginosa 5086 (PA5086) | | Descriptor: | Sel1 repeat family protein | | Authors: | She, Z, Geng, Z. | | Deposit date: | 2019-11-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa T6SS PldB immunity proteins PA5086, PA5087 and PA5088 explains a novel stockpiling mechanism.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8IVX
 
 | | Crystal structure of NRP2 in complex with aNRP2-14 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody 14V4 Fab fragment, Light chain of antibody 14V4 Fab fragment, ... | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
2JP5
 
 | | ATWLPPR an anti-angiogenic peptide | | Descriptor: | ATWLPPR peptide | | Authors: | Badache, S, Bouchemal, N.C, Herve du Penhoat, C.L.M. | | Deposit date: | 2007-04-20 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the antiangiogenic ATWLPPR peptide inhibiting VEGF(165) binding to neuropilin-1 and molecular dynamics simulations of the ATWLPPR/neuropilin-1 complex
Peptides, 28, 2007
|
|
7KBB
 
 | |
7KBA
 
 | |
6GH8
 
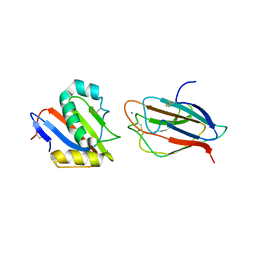 | | Crystal structure of GP1 domain of Lujo virus in complex with the first CUB domain of neuropilin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glycoprotein, ... | | Authors: | Cohen-Dvashi, H, Kilimnik, I, Diskin, R. | | Deposit date: | 2018-05-06 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for receptor recognition by Lujo virus.
Nat Microbiol, 3, 2018
|
|
5TTD
 
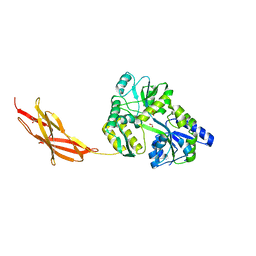 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
7M1C
 
 | |
7LYV
 
 | |
7LYW
 
 | |
