3FK0
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FJX
 
 | | E. coli EPSP synthase (T97I) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FJZ
 
 | | E. coli EPSP synthase (T97I) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3FK1
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
9FL3
 
 | | Crystal structure of IL-17A in complex with compound 26 | | Descriptor: | (~{E})-~{N}-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[7-[(1~{S})-2-methoxy-1-[(4~{S})-2-oxidanylidene-4-(trifluoromethyl)imidazolidin-1-yl]ethyl]imidazo[1,2-b]pyridazin-2-yl]methyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 67, 2024
|
|
2XG5
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 5d | | Descriptor: | (2R)-2-[5-CYCLOPROPYL-6-(HYDROXYSULFANYL)-4-(NAPHTHALEN-1-YLMETHYL)-2-OXOPYRIDIN-1(2H)-YL]-3-PHENYLPROPANOIC ACID, (2R,3R)-8-CYCLOPROPYL-7-(NAPHTHALEN-1-YLMETHYL)-5-OXO-2-PHENYL-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
5WSN
 
 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
2XG4
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, COBALT (II) ION, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
5MKJ
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 9 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(5-prop-2-enyl-2-propoxy-phenyl)phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
5MKU
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
2W0Q
 
 | | E. coli copper amine oxidase in complex with Xenon | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE, ... | | Authors: | Pirrat, P, Smith, M.A, Pearson, A.R, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1DP0
 
 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
6SCZ
 
 | | Mycobacterium tuberculosis alanine racemase inhibited by DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Chiara, C, Purkiss, A, Prosser, G, Homsak, M, de Carvalho, L.P.S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition.
Nat.Chem.Biol., 16, 2020
|
|
6RB1
 
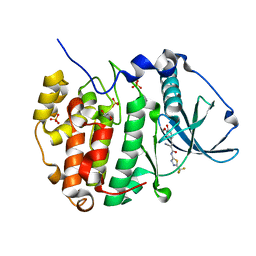 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 1 | | Descriptor: | (~{E})-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)-~{N}-[5-(trifluoromethyl)-1,3,4-thiadiazol-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
6RCB
 
 | |
6RGS
 
 | |
5OR6
 
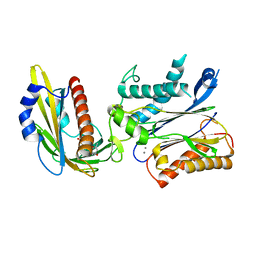 | | Crystal structures of PYR1/HAB1 in complex with synthetic analogues of Abscisic Acid | | Descriptor: | (~{E})-3-(trifluoromethyl)-5-[(1~{S})-2,6,6-trimethyl-1-oxidanyl-4-oxidanylidene-cyclohex-2-en-1-yl]pent-2-en-4-ynoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the in Vitro and in Vivo SAR of Abscisic Acid - Exploring Unprecedented Variations of the Side Chain via Cross-Coupling-Mediated Syntheses
Eur.J.Org.Chem., 2018
|
|
6AEJ
 
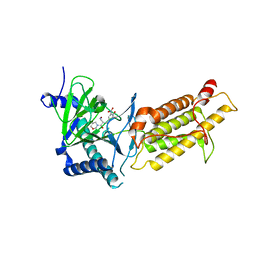 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (E)-3-[3-nitro-4,5-bis(oxidanyl)phenyl]-2-(1,3-oxazinan-3-ylcarbonyl)prop-2-enenitrile, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
3I3D
 
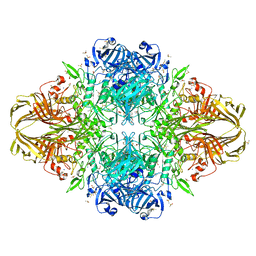 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
1F4H
 
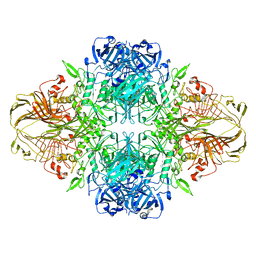 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
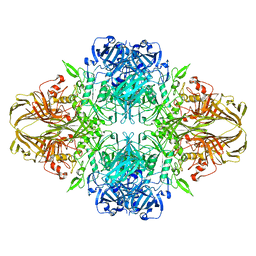 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
3I3E
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
3I3B
 
 | | E.coli (lacz) Beta-Galactosidase (M542A) in Complex with D-Galactopyranosyl-1-on | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
5MMW
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 6 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-3-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6AZ1
 
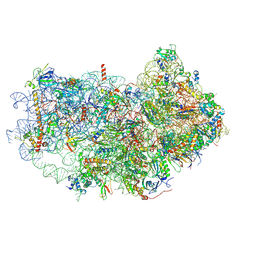 | | Cryo-EM structure of the small subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | E-site tRNA, LACK1, MAGNESIUM ION, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Matzov, D, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
