5T35
 
 | | The PROTAC MZ1 in complex with the second bromodomain of Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Transcription elongation factor B polypeptide 1, ... | | Authors: | Gadd, M.S, Zengerle, M, Ciulli, A. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of PROTAC cooperative recognition for selective protein degradation.
Nat. Chem. Biol., 13, 2017
|
|
5V1Y
 
 | | Crystal structure of the ternary RPN13 PRU-RPN2 (940-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
4II3
 
 | |
5U4P
 
 | | Protein-protein complex between 26S proteasome regulatory subunit RPN8, RPN11, and Ubiquitin S31 | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8, Ubiquitin-40S ribosomal protein S31, ... | | Authors: | Worden, E.J, Dong, K.C, Martin, A. | | Deposit date: | 2016-12-05 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome.
Mol. Cell, 67, 2017
|
|
4JQW
 
 | | Crystal Structure of a Complex of NOD1 CARD and Ubiquitin | | Descriptor: | Nucleotide-binding oligomerization domain-containing protein 1, PHOSPHATE ION, Polyubiquitin-C | | Authors: | Ver Heul, A.M, Gakhar, L, Piper, R.C, Ramaswamy, S. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a complex of NOD1 CARD and ubiquitin
Plos One, 9, 2014
|
|
4JIO
 
 | | Bro1 V domain and ubiquitin | | Descriptor: | BRO1, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Piper, R.C. | | Deposit date: | 2013-03-06 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The yeast alix homolog bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes.
Dev.Cell, 25, 2013
|
|
5V1Z
 
 | | Crystal structure of the RPN13 PRU-RPN2 (932-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
4K1R
 
 | |
5HPS
 
 | |
5HPK
 
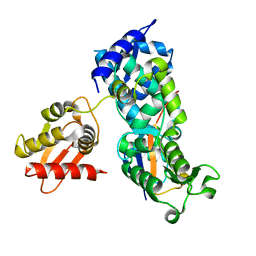 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
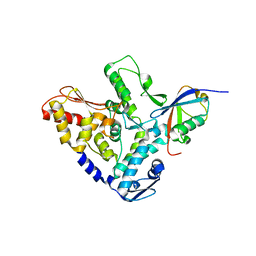
5HPL
 
 | |
2DX5
 
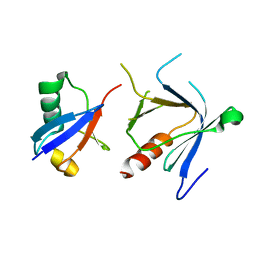 | | The complex structure between the mouse EAP45-GLUE domain and ubiquitin | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Hirano, S, Suzuki, N, Slagsvold, T, Kawasaki, M, Trambaiolo, D, Kato, R, Stenmark, H, Wakatsuki, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Nat.Struct.Mol.Biol., 13, 2006
|
|
5J26
 
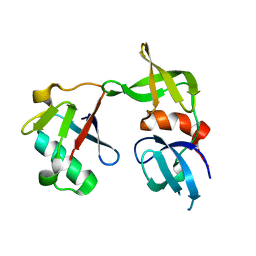 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
5JG6
 
 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5IBK
 
 | | Skp1-F-box in complex with a ubiquitin variant | | Descriptor: | F-box/WD repeat-containing protein 7, Polyubiquitin-B, S-phase kinase-associated protein 1,S-phase kinase-associated protein 1 | | Authors: | Orlicky, S, Sicheri, F. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
2C7N
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
5JP3
 
 | |
6ML1
 
 | | Structure of the USP15 deubiquitinase domain in complex with an affinity-matured inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
5JQS
 
 | |
5JTJ
 
 | | USP7CD-CTP in complex with Ubiquitin | | Descriptor: | CALCIUM ION, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7,Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5JTV
 
 | | USP7CD-UBL45 in complex with Ubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
6N6R
 
 | |
6N5M
 
 | |
2C7M
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin.
Cell, 124, 2006
|
|
