3VD8
 
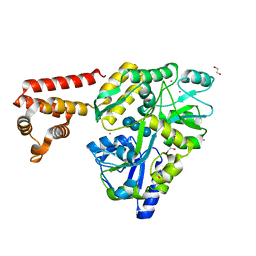 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
6KXG
 
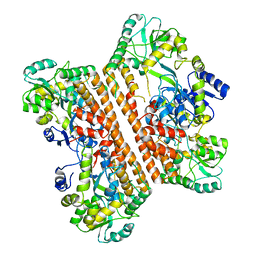 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
6LES
 
 | |
6EQZ
 
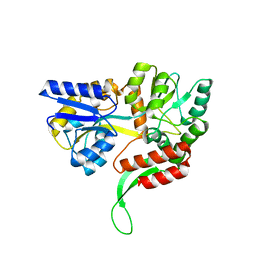 | | A MamC-MIC insertion in MBP scaffold at position K170 | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Zarivach, R. | | Deposit date: | 2017-10-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6OB5
 
 | | Computationally-designed, modular sense/response system (S3-2D) | | Descriptor: | Ankyrin Repeat Domain (AR), S3-2D variant, FARNESYL DIPHOSPHATE, ... | | Authors: | Thompson, M.C, Glasgow, A.A, Huang, Y.M, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Computational design of a modular protein sense-response system.
Science, 366, 2019
|
|
6EG3
 
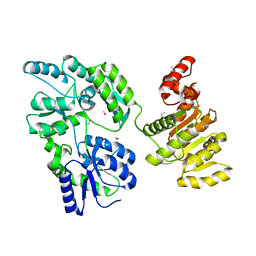 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6O7H
 
 | |
3VFJ
 
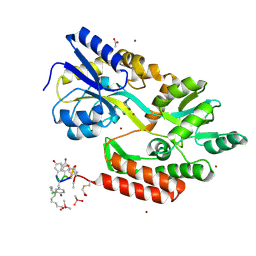 | | The structure of monodechloro-teicoplanin in complex with its ligand, using MBP as a ligand carrier | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Loll, P.J. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the complex between teicoplanin and a bacterial cell-wall peptide: use of a carrier-protein approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6EG2
 
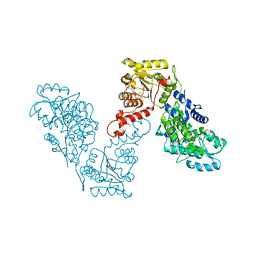 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6USM
 
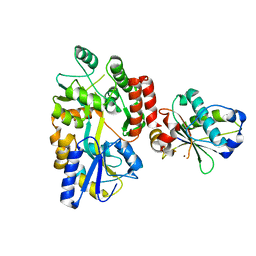 | |
6OFW
 
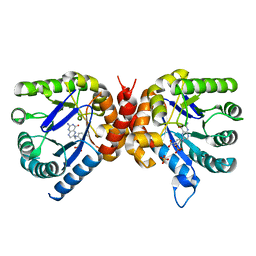 | | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate | | Descriptor: | 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}benzoic acid, Dihydropteroate synthase, SULFATE ION | | Authors: | Oliveira, A.A, Guido, R.V.C, Maluf, F.V, Bueno, R.V, Lima, G.M.A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dihydropteroate synthase from Xanthomonas albilineans in complex with 7,8-dihydropteroate
To Be Published
|
|
3A3C
 
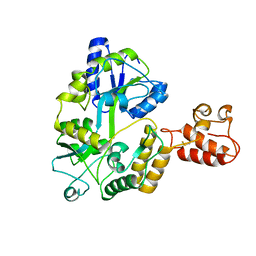 | | Crystal structure of TIM40/MIA40 fusing MBP, C296S and C298S mutant | | Descriptor: | Maltose-binding periplasmic protein, LINKER, Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Kawano, S, Naoe, M, Momose, T, Watanabe, N, Endo, T. | | Deposit date: | 2009-06-11 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3W15
 
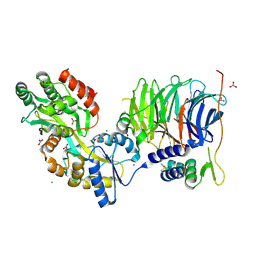 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
6V3E
 
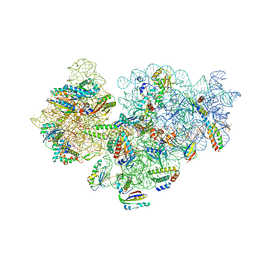 | |
6P6W
 
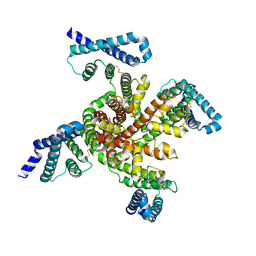 | | Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant in the resting state | | Descriptor: | Fusion of Maltose-binding protein and voltage-gated sodium channel NavAb | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-Din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
6FCL
 
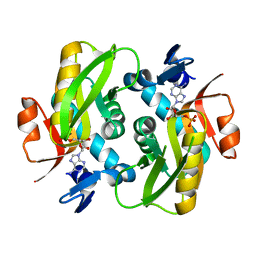 | |
6FD6
 
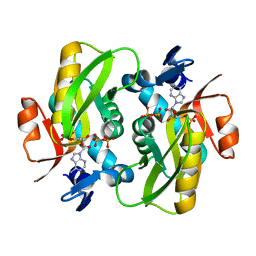 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 30 days post crystallization (with AMP and PPi products fully generated) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
6PMJ
 
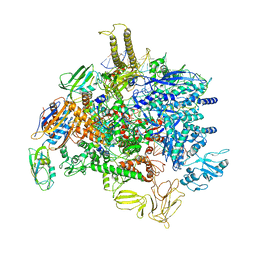 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6V2E
 
 | |
6V39
 
 | |
3WJO
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with isopentenyl pyrophosphate (IPP) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
2ZXT
 
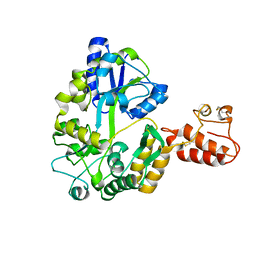 | | Crystal structure of Tim40/MIA40, a disulfide relay system in mitochondria, solved as MBP fusion protein | | Descriptor: | Maltose-binding periplasmic protein, LINKER, Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Kawano, S, Momose, T, Watanabe, N, Endo, T. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3WJK
 
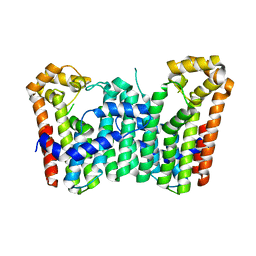 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli | | Descriptor: | Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
6PGQ
 
 | |
3BJI
 
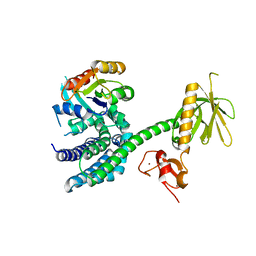 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
