1ASU
 
 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 2% PEG 400, 2M AMMONIUM SULFATE, HEPES PH 7.5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
4YUD
 
 | |
9NCW
 
 | | Crystal Structure of WDR5 in complex with Triazole-Based Inhibitors | | Descriptor: | 1-{[(6P)-3-[(3,5-dimethoxyphenyl)methyl]-6-(4-fluoro-2-methylphenyl)-4,5-dihydro-3H-naphtho[1,2-d][1,2,3]triazol-8-yl]methyl}-3-methyl-1,3-dihydro-2H-imidazol-2-imine, WD repeat-containing protein 5 | | Authors: | Goins, C.M, Stauffer, S.R. | | Deposit date: | 2025-02-17 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Development and Characterization of Triazole-Based WDR5 Inhibitors for the Treatment of Glioblastoma
Biorxiv, 2025
|
|
1ZKD
 
 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
5U0R
 
 | |
6EP8
 
 | | InhA Y158F mutant in complex with NADH from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Wagner, T, Voegeli, B, Rosenthal, R.G, Stoffel, G, Shima, S, Kiefer, P, Cortina, N, Erb, T.J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | InhA, the enoyl-thioester reductase fromMycobacterium tuberculosisforms a covalent adduct during catalysis.
J. Biol. Chem., 293, 2018
|
|
2WPK
 
 | | factor IXa superactive triple mutant, ethylene glycol-soaked | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
5YWN
 
 | | SsCR_L211H-NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
7JTW
 
 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | Descriptor: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2020-08-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
5XDL
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Zhu, S.J, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
7L34
 
 | | Human DNA Ligase 1 - R641L nicked DNA complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Tumbale, P.P, Williams, R.S, Schellenberg, M.S. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | LIG1 syndrome mutations remodel a cooperative network of ligand binding interactions to compromise ligation efficiency.
Nucleic Acids Res., 49, 2021
|
|
7JNT
 
 | |
5WZM
 
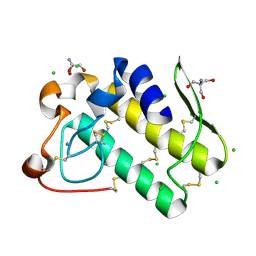 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
6GA5
 
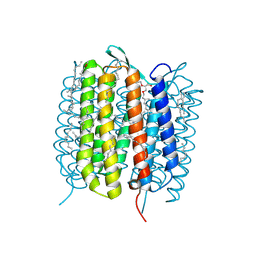 | | Bacteriorhodopsin, 3 ps state, REAL-SPACE REFINEMED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5X26
 
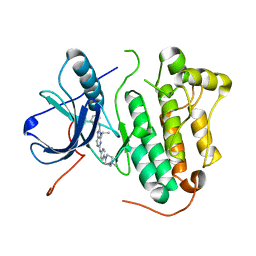 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
3NSZ
 
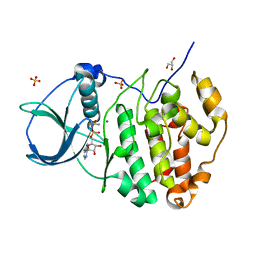 | | Human CK2 catalytic domain in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
7ZLD
 
 | | Crystal Structure of Unlinked NS2B_NS3 Protease from Zika Virus in Complex with Inhibitor MI-2223 | | Descriptor: | (2~{S})-2-[2-[3-(aminomethyl)phenyl]ethanoylamino]-6-azanyl-~{N}-[(2~{S})-6-azanyl-1-[[(5~{R})-6-azanyl-5-carbamimidamido-6-oxidanylidene-hexyl]amino]-1-oxidanylidene-hexan-2-yl]hexanamide, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Thermodynamic characterization of a macrocyclic Zika virus NS2B/NS3 protease inhibitor and its acyclic analogs.
Arch Pharm, 356, 2023
|
|
7ZIQ
 
 | | BK Polyomavirus VP1 in complex with 6'-Sialyllactose glycomacromolecules (aromatic linker) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
6TRI
 
 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5X2K
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with WZ4003 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Zhu, S.J, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
7ZLC
 
 | |
7ZZA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | 2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-[(3-azanylpropylcarbamoylamino)methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]ethanoic acid, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
6G2I
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) at 5.9 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1, Breast cancer type 1 susceptibility protein | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
4UOW
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
