7W7B
 
 | | Heme exporter HrtBA in complex with protoporphyrin IX containing manganese(III), high resolution data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7C
 
 | | Heme exporter in the unliganded form | | Descriptor: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | Authors: | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7E
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7QER
 
 | | human Connexin 26 dodecamer at 55mm Hg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEQ
 
 | | human Connexin 26 dodecamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QET
 
 | | human Connexin 26 dodecamer at 20mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEW
 
 | | human Connexin 26 class 2 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEY
 
 | | human Connexin 26 class 1 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEU
 
 | | human Connexin 26 at 55mmHg PCO2, pH7.4: two masked subunits, class B | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEV
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4:two masked subunits, class D | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEO
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QES
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class A | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7T1K
 
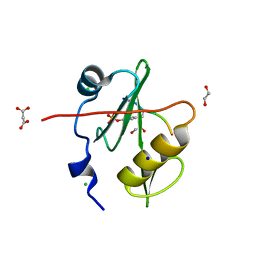 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1L
 
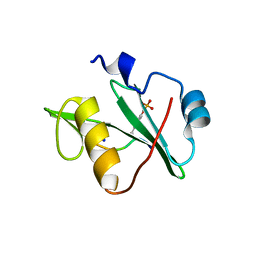 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1U
 
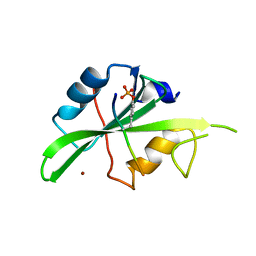 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
7W6P
 
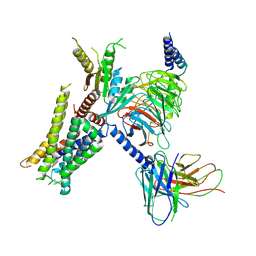 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W6W
 
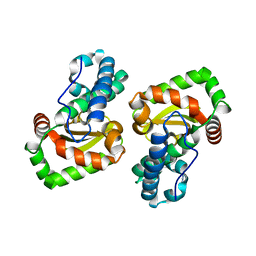 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase L169W | | Descriptor: | AZIDE ION, MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7T1C
 
 | |
7T1F
 
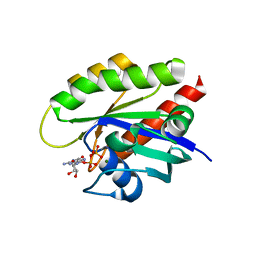 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
7T1E
 
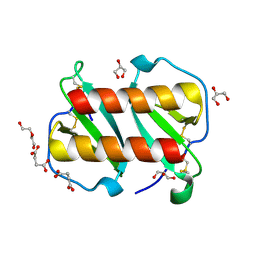 | | Structure of monomeric and dimeric human CCL20 | | Descriptor: | C-C motif chemokine 20, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Peterson, F.C, Riutta, S.J, Volkman, B.F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | The Chemokine, CCL20, and Its Receptor, CCR6, in the Pathogenesis and Treatment of Psoriasis and Psoriatic Arthritis
J Psoriasis Psoriatic Arthritis, 8, 2023
|
|
7QE8
 
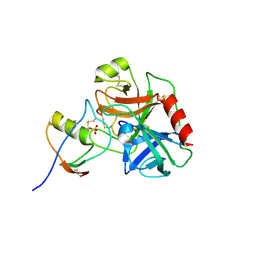 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 (SPINK1) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7QE9
 
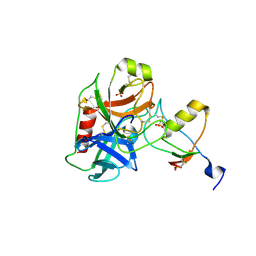 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 N34S (SPINK1 N34S) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7W5O
 
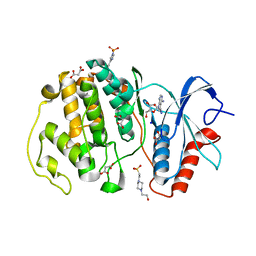 | | Crystal structure of ERK2 with an allosteric inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a novel target site for ATP-independent ERK2 inhibitors.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7T0O
 
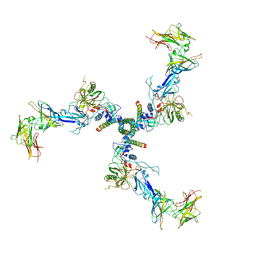 | |
