7C2N
 
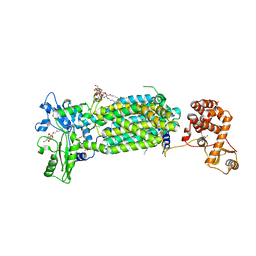 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
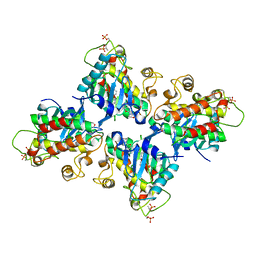
7C2W
 
 | |
5F3A
 
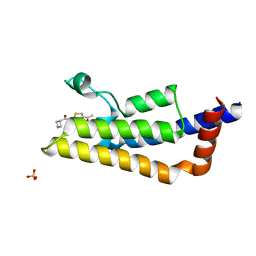 | |
5B5B
 
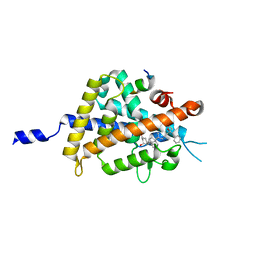 | | Crystal structure of VDR-LBD complexed with 2-methylidene-26,27-diphenyl-19-nor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-oxidanyl-7-phenyl-6-(phenylmethyl)heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix12-Stabilization Antagonist of Vitamin D Receptor
Bioconjug.Chem., 27, 2016
|
|
6KBY
 
 | | Crystal structure of a class C beta lactamase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
7BTH
 
 | | Mevo lectin- Native form-1 | | Descriptor: | GLYCEROL, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
3MNR
 
 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
5AYC
 
 | | Crystal structure of Ruminococcus albus 4-O-beta-D-mannosyl-D-glucose phosphorylase (RaMP1) in complexes with sulfate and 4-O-beta-D-mannosyl-D-glucose | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, SULFATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
6A5Y
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC143 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
1ZJ5
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6Q9I
 
 | |
6SXV
 
 | | GH51 a-l-arabinofuranosidase soaked with aziridine inhibitor | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GH51 a-l-arabinofuranosidase, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
6DDJ
 
 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
4BOW
 
 | | Crystal structure of LamA_E269S from Z. galactanivorans in complex with laminaritriose and laminaritetraose | | Descriptor: | CALCIUM ION, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Beta-Glucanase Zglama from Zobellia Galactanivorans Evolved a Bent Active Site Adapted for Efficient Degradation of Algal Laminarin
J.Biol.Chem., 289, 2014
|
|
4Y37
 
 | | Endothiapepsin in complex with fragement 305 | | Descriptor: | 4-chloro-2-methylthieno[2,3-d][1,2,3]diazaborinin-1(2H)-ol, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5ZRI
 
 | | M. smegmatis antimutator protein MutT2 in complex with 5m-dCMP | | Descriptor: | MAGNESIUM ION, Putative mutator protein MutT2/NUDIX hydrolase, [(2R,3S,5R)-5-(4-azanyl-5-methyl-pyrimidin-1-ium-1-yl)-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
7QH9
 
 | | TarM(Se)_G117R-4RboP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TarM(Se)_G117R-4RboP, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5EZP
 
 | | Human transthyretin (TTR) complexed with 4-hydroxy-chalcone | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-chalcone, Transthyretin | | Authors: | Polsinelli, I, Nencetti, S, Shepard, W.E, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new crystal form of human transthyretin obtained with a curcumin derived ligand.
J.Struct.Biol., 194, 2016
|
|
6STG
 
 | | Human Rab8a phosphorylated at Ser111 in complex with GPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vieweg, S, Mulholland, K, Braeuning, B, Kachariya, N, Lai, Y, Toth, R, Sattler, M, Groll, M, Itzen, A, Muqit, M.M.K. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72.
Biochem.J., 477, 2020
|
|
6KMM
 
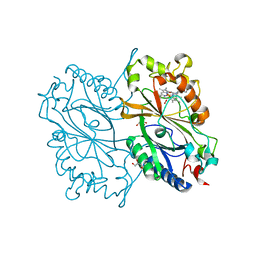 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
4C2I
 
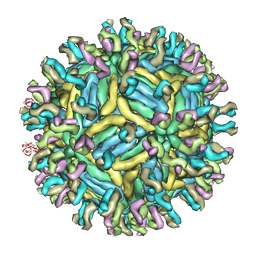 | | Cryo-EM structure of Dengue virus serotype 1 complexed with Fab fragments of human antibody 1F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fibriansah, G, Tan, J.L, de Alwis, R, Smith, S.A, Ng, T.-S, Kostyuchenko, V.A, Ibarra, K.D, Harris, E, de Silva, A, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2013-08-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A Potent Anti-Dengue Human Antibody Preferentially Recognizes the Conformation of E Protein Monomers Assembled on the Virus Surface.
Embo Mol.Med., 6, 2014
|
|
5ON6
 
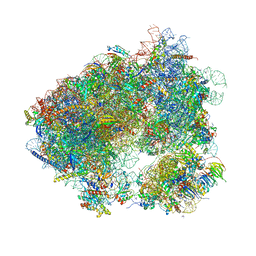 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
7XMH
 
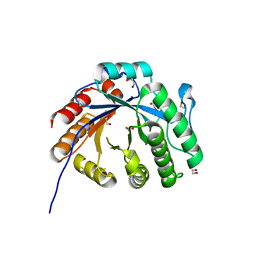 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
